This chapter deals with the biology of rice (Oryza sativa), a revision of the original 1999 publication. It contains information for use during the risk/safety regulatory assessment of genetically engineered varieties of rice intended to be grown in the environment (biosafety). It includes elements of taxonomy, centres of origin, cultivation, reproductive biology, genetics, hybridisation and introgression, as well as ecology. Annexes present a glossary of rice ecological types, the common diseases, pests and weeds in rice fields, and current biotechnology developments.
Safety Assessment of Transgenic Organisms in the Environment, Volume 9

4. Biology of Rice (Oryza sativa)
Abstract
Introduction
This chapter was prepared by the OECD Working Party on the Harmonisation of Regulatory Oversight in Biotechnology, with Japan as the lead country. It was initially issued in 2021 as the Revised Consensus Document on the Biology of Rice (Oryza sativa L.), replacing the original document issued in 1999. Production and trading data have been updated in this publication, based on FAOSTAT.
General description including taxonomy and morphology
Classification and nomenclature
Rice is the common name for all plant members belonging to the genus Oryza in the Poaceae (Gramineae) family. Cultivated rice includes two species: Oryza sativa L. and Oryza glaberrima Steud. Scientifically, wild rice refers to all other Oryza species, excluding the two cultivated species. However, when commonly used, the term wild rice additionally includes both cultivated and wild species from the genus Zizania and Porteresia, which are closely related but do not belong to the genus Oryza.
The cultivated rice species O. sativa is classified into two sub-species, indica and japonica, which are roughly characterised as long-grain rice and short-grain rice respectively (Figure 4.1). Japonica rice has been further classified into two ecotypes, temperate and tropical japonica according to the ecosystems where they have been evolved, whereas indica rice has been classified into ecotypes such as Aus, Amman, Rayada, and Boro according to their growing time and locations (Morinaga, 1968). Analysis by Wang et al. (2018) using genome sequencing of 3 010 accessions has revealed that O. sativa can be classified into nine sub-populations: four sub-populations for indica, three sub-populations for japonica and two single groups. These nine sub-populations could be related to geographic location and all these accessions are collectively called Asian cultivated rice. These Asian cultivated rice varieties have been widely grown in Asia, Oceania, Africa, North, Central, and South America, and Europe. More than 80% of these are represented by indica rice and approximately 15% by japonica rice. Another cultivated species, O. glaberrima, which is grown in limited areas in Africa, accounts for a small percentage (less than 5%) of global rice production and is known as African cultivated rice.
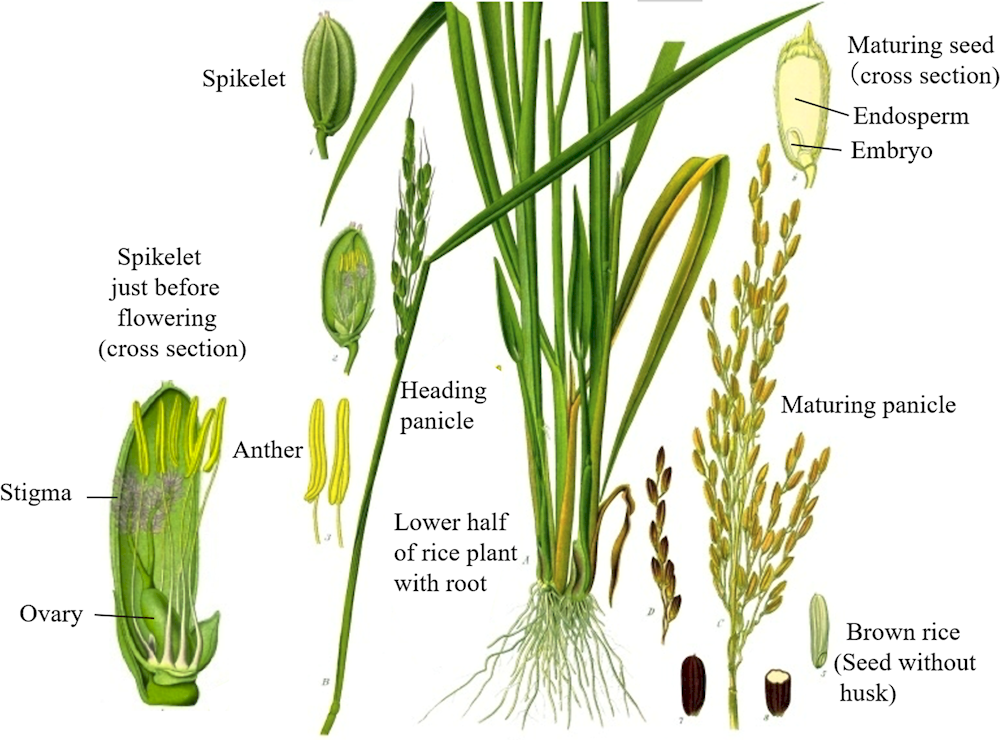
Figure 4.1. Panicle, seeds and brown rice of typical cultivated rice

Note: Indica rice (left), japonica rice (right).
Source: a) Photo zou (accessed 22 March 2022), http://photozou.jp/; b, c) courtesy of NARO; d) photo AC (accessed 22 March 2022), https://premium.photo-ac.com/; e, f) NARO (accessed 22 March 2022), Genebank Project, https://www.gene.affrc.go.jp/databases.php.
The genus Oryza comprises 23 species, including cultivated and wild species, and is divided into four major complexes (Table 4.1): the sativa complex with eight species, the officinalis complex, which is the largest with 11 species, the ridleyanae complex and the granulata complex, the latter two being remote relatives of O. sativa (Wing, Purugganan and Zhang, 2018; Zou et al., 2015; Joseph, Kuriachan and Thomas, 2008). Asian cultivated rice is thought to originate from an ancestor of perennial wild rice O. rufipogon, via multiple crossings between evolving plants that ultimately differentiated into the presently known species, sub‑species and varieties (Chen et al., 2019; Choi et al., 2019). Another cultivated rice species, O. glaberrima, was found to have evolved from wild rice O. barthii (Wang et al., 2014). The habitats of eight species in the sativa complex, two cultivated species (O. sativa and O. glaberrima) and six close wild species relatives are shown in Figure 4.2.
Table 4.1. Classification and distribution of 23 species in the genus Oryza
|
Class |
Species |
Chromosome number |
Genome |
Genome size1 |
Genome size2 |
Distribution |
|---|---|---|---|---|---|---|
|
Section sativa |
Oryza sativa L. |
24 |
AA |
390 |
Asia |
|
|
O. rufipogon sensu lato |
24 |
AA |
439 |
381 |
Asia, Oceania |
|
|
O. nivara |
24 |
AA |
448 |
Asia, Oceania |
||
|
O. glaberrima Steud. |
24 |
AA |
3573 |
West Africa |
||
|
O. barthii A. Chev. |
24 |
AA |
403 |
Africa |
||
|
O. longistaminata Chev. & Roehr |
24 |
AA |
390 |
Africa |
||
|
O. meridionalis Ng |
24 |
AA |
381 |
Australia |
||
|
O. glumaepatula Steud. |
24 |
AA |
437 |
Central and South America |
||
|
Section officinalis |
O. officinalis Wall ex Watt |
24 |
CC |
651 |
549 |
Asia |
|
O. rhizomatis D.A. Vaughan |
24 |
CC |
823 |
Sri Lanka |
||
|
O. eichingeri Peter |
24 |
CC |
587 |
Africa, Sri Lanka |
||
|
O. minuta J.S. Presl ex C.B. Presl. |
48 |
BBCC |
1 1243 |
Philippines |
||
|
O. punctata Kotechy ex Steud. |
24, 48 |
BB,BBCC |
425 |
364 |
Africa |
|
|
O. latifolia Desv. |
48 |
CCDD |
806 |
Central and South America |
||
|
O. alta Swallen |
48 |
CCDD |
1 008 |
866 |
Central and South America |
|
|
O. grandiglumis (Döll.) Prod |
48 |
CCDD |
891 |
South America |
||
|
O. australiensis Domin |
24 |
EE |
965 |
827 |
Australia |
|
|
Section ridleyanae |
O. brachyantha Chev. & Roehr. |
24 |
FF |
362 |
265 |
Africa |
|
O. ridleyi Hook |
48 |
HHJJ |
1 283 |
1 080 |
Asia, New Guinea |
|
|
O. longiglumis Jansen |
48 |
HHJJ |
1 209 |
New Guinea |
||
|
O. schlechteri Pilger |
48 |
unknown |
Papua New Guinea |
|||
|
Section granulata |
O. granulata Nees & Am ex Watt |
24 |
GG |
882 |
1 016 |
Asia |
|
O. meyeriana (Zoll. & Mor.ex Steud.) Baill |
24 |
GG |
1 003 |
Asia |
1. Ammiraju, J.S.S. et al. (2006), “The Oryza bacterial artificial chromosome library resources: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza”, https://dx.doi.org/10.1101%2Fgr.3766306.
2. Calculated from Miyabayashi, T. et al. (2007), “Genome size of twenty wild species of Oryza Species determined by flow cytometric and chromosome analyses”, https://doi.org/10.1270/jsbbs.57.73.
3. Martínez, C.P. et al. (1994), “Nuclear DNA content of ten rice species as determined by flow cytometry”, https://doi.org/10.1266/jjg.69.513.
Other sources: Wing, R., M.D. Purugganan and Q. Zhang (2018), “The rice genome revolution: From an ancient grain to Green Super Rice”, https://doi.org/10.1038/s41576-018-0024-z; Zou, X.H. et al. (2015), “Multiple origins of BBCC allopolyploid species in the rice genus (Oryza)”, https://doi.org/10.1038/srep14876; Joseph, L., P. Kuriachan and G. Thomas, (2008), “Is Oryza malampuzhaensis Krish. et Chand. (Poaceae) a valid species? Evidence from morphological and molecular analyses”, https://doi.org/10.1007/s00606-007-0606-2.
Figure 4.2. Habitats of eight species of the sativa complex in the genus Oryza
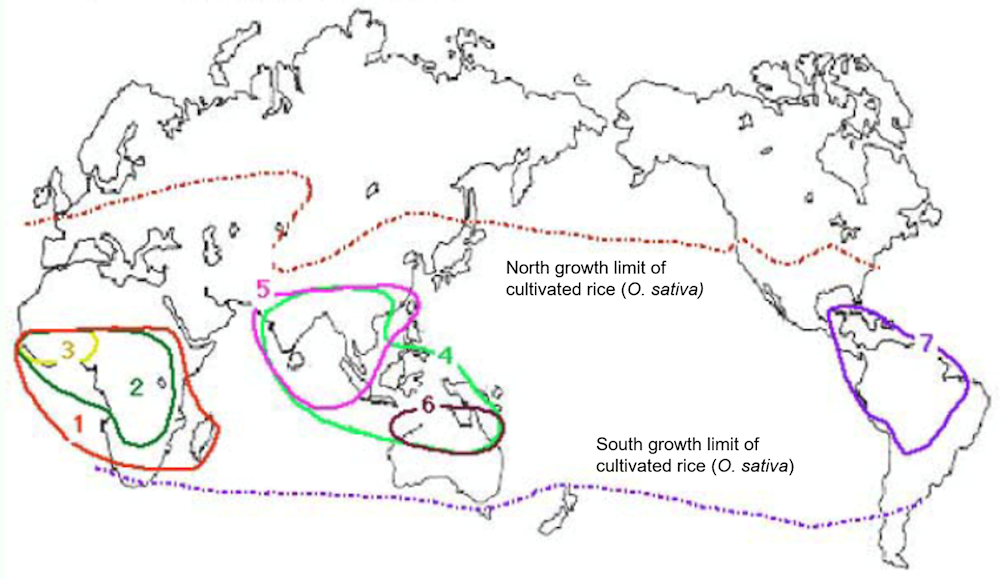
Note: 1. O. longistaminata; 2. O. barthii; 3. O. glaberrima; 4. O. rufipogon (perennial); 5. O. nivara (annual O. rufipogon); 6. O. meridionalis; 7. O. glumaepatula.
Source: Adapted from Stein, J.C. et al. (2018), “Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza”, https://doi.org/10.1038/s41588-018-0040-0, and Oryzabase https://shigen.nig.ac.jp/rice/oryzabase/.
It has been suggested that Asian cultivated rice originated in the lower valley of the Yangtze River, People’s Republic of China (hereafter ‘China’), based on geographical, archaeological and folkloric traces (Fuller et al., 2009). However, more recent molecular biological analysis using a number of living rice accessions has indicated that the point of origin may be in the lower valley of Pearl River (Huang et al., 2012). Several hypotheses have been proposed regarding rice cultivation events, such as single cultivation events, independent multiple cultivation events and single cultivation followed by multiple crossing events. As of 2020, the results from the genome analyses of a large number of Asian cultivated and wild rice varieties/accessions support the hypothesis that multiple cultivated sub-species and varieties differentiated from a single cultivated variety, which was crossed with multiple wild/cultivated accessions. Details are also shown in the subsection on centres of origin and diversity, and geographical distribution.
Plants in the genus Oryza have a basis of 12 chromosomes, both in diploid species with two sets (2n = 24) and allotetraploid species with four sets (2n = 48). Each chromosome set is designated with 1 of the following 11 genome names: A, B, C, D, E, F, G, H, J, K or L, according to the chromosome pairing affinity of hybrid plants (Ge et al., 1999) or comparisons of the genome sequences (Wing, Purugganan and Zhang, 2018). The most popular species O. sativa was the first categorised as the “A” genome; subsequently, unknown species were given their alphabetical genome names in order of discovery (Kurata and Omura, 1984; Wing, Purugganan and Zhang, 2018). Genome size was found to vary between species ranging from 265 megabases (Mb) (O. brachyantha, an F genome species) to 827 Mb (O. australiensis, an E genome species), and the reference genome of O. sativa, japonica was 389 Mb (Miyabayashi et al., 2007). Table 4.1 summarises the chromosome number, ploidy level, genome name, genome size and habitats for all Oryza species.
The evolutionary relationships among all Oryza species have been clarified in previous studies (Figure 4.3) (Wing, Purugganan and Zhang, 2018). O. granulata emerged first approximately 14 million years ago (mya) and the AA genome species, including cultivated species, evolved most recently, approximately 3 mya (Stein et al., 2018). The O. sativa subsp. japonica rice genome was fully sequenced in 2004 by the International Rice Genome Sequencing Project (2005) and successive comparative genomic studies have been carried out for other species and genomes. These studies have found that genome construction appears to be similar among the different Oryza species, with genome-long homology and gene order conservation, while most of the variation in genome size was derived from amplification and defects of various types of transposable elements and from the copy numbers of specific gene families (Copetti and Wing, 2016; Wing, Purugganan and Zhang, 2018).
Figure 4.3. Evolutionary relationships of species in the genus Oryza
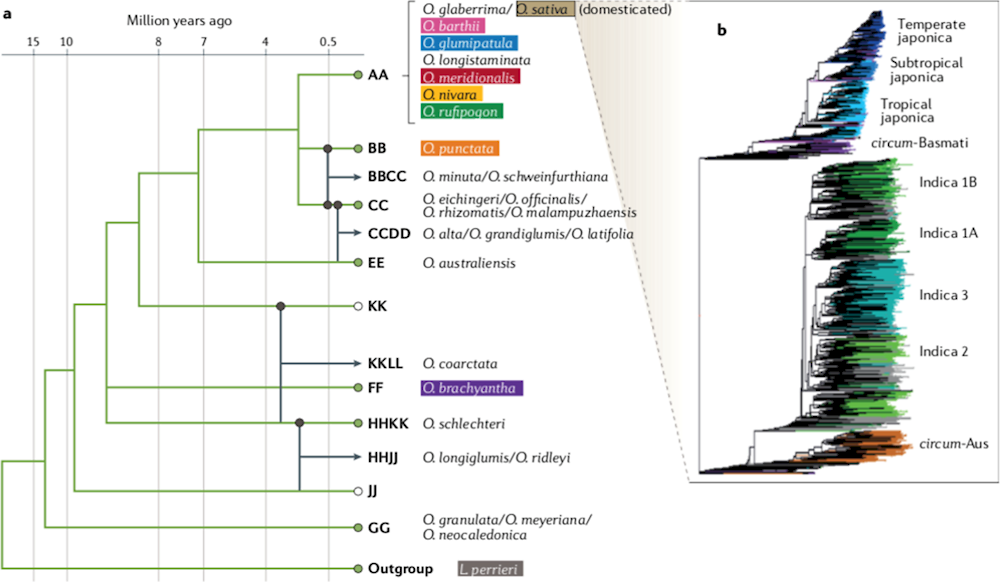
Source: Wing, R., M.D. Purugganan and Q. Zhang (2018), “The rice genome revolution: From an ancient grain to Green Super Rice”, https://doi.org/10.1038/s41576-018-0024-z.
Asian cultivated rice O. sativa propagates by self-pollination, with up to 6.8% intraspecies outcrossing and cross-fertilisation with other species occurring at various rates. O. rufipogon and O. nivara, which are the closest relatives of O. sativa, have high cross-fertilisation abilities with O. sativa as they can easily cross with each other to yield offspring. Therefore, in habitats where the cultivated varieties and wild species overlap, weedy rice, which is derived from crosses between cultivated and wild rice, grows wildly and is problematic. Other AA genome species aside from these three closely‑related species have relatively lower cross-fertilisation abilities and can yield hybrid embryos when crossed with the cultivated rice O. sativa. However, the embryos from such genetic combinations have various difficulties surviving because of the reproductive barriers that exist in their seed formation and/or hybrid plant growth. Meanwhile, the crossing of cultivated rice with wild Oryza species other than AA genome species is much more difficult and fertile seeds are rarely propagated.
In addition to the rice categories for the sub-species and ecotypes mentioned above, when considering the interactions between rice and other organisms (discussed in the section “Various interactions with other organisms (ecology)”), rice is also categorised into several ecological types: cultivated, volunteer, weedy, and wild rice types. The characteristics of each type and relationships among these types are described in Annex 4.A. These rice types can easily cross with each other, produce offspring and grow on and off farms as hybrid swarms. They consequently sometimes become problematic for rice farming.
Description
Cultivated rice is a herbaceous annual crop with an erect habit. Under standard cultivation conditions, the plant height ranges from 0.5 to 2 metres. The leaf blade ranges from 1 to 3 cm wide and from 20 to 50 cm long and is connected to the leaf sheath by a collar consisting of both the ligule and auricle. Tiller (culm) development is closely synchronised with leaf emergence in the upper three nodes on the corresponding stem; a total of 5-20 tillers per plant emerge, and the number depends on the total growth duration and planting density.
Under standard cultivation, floral development followed by panicle initiation starts 50 to 100 days after germination. The panicle or inflorescence emerges from the top of the culm after an additional 25-30 days and this event is defined as heading. The panicle is conical and its length ranges from 10 to 40 cm. Under normal growth, it sets from 50 to 400 spikelets on 10 to 20 branches. Each spikelet has six stamens (anthers) and one pair of pistils (i.e. two stigmas).
At the time of flowering, on the morning of a sunny summer’s day, 6 000‑12 000 pollen grains are dispersed from the split anthers and attach to the stigmas, resulting in fertilisation. During the 30 days after fertilisation, the embryo and endosperm develop into mature rice grains, which become seed for the propagation of the next generation.
Edible brown rice is yielded by removing the husk from the grain, while white rice is yielded by then removing the bran layers and embryo (germ) from the brown rice, leaving only the endosperm. The morphological characteristics of a typical rice plant are shown in Figure 4.4.
Figure 4.4. Phenotypic characteristics of cultivated rice O. sativa
The weight of fully matured seeds ranges from 10 to 46 mg (mode, 22-25 mg) among all varieties. The ratio of seed length to width varies widely from 1.6:1 for round grains to 4.0:1 for long grains. Generally, seeds retain dormancy traits that inhibit germination immediately after harvest. The degree of seed dormancy differs depending on the cultivar and environmental conditions, even though it is usually lost after a year in normal conditions (20-30°C). Dormancy can be coercively cancelled by heat treatment, hydrochloric acid treatment or the removal of the husk. If dormancy is broken, seeds start germination when their water content exceeds 30% after 24-48 hours of being soaked in water at 30°C. Seeds can maintain their germination ability for more than 10 years if they are kept at low temperatures and under dry conditions (less than 10% humidity).
The life cycle and morphological characteristics described above are typical of average rice plants grown around the world but they fluctuate greatly, even for the same cultivar, depending on the soil and weather conditions. Additionally, wild rice, mutant rice or cultivars grown in specific environments can exhibit extreme phenotypes deviating from the values described here.
Based on the starch content and utilisation of the edible grain, rice can be classified as glutinous or non‑glutinous. Each of these types is further categorised by length as long grain or short grain, by fragrance (fragrant or not) and by colour (white, red, purple or black). According to these classifications, rice is processed in various styles such as boiled, stir-fried or steamed, for eating. Rice flour is kneaded and steamed or baked to produce various processed foods, such as noodles and cookies. Rice bran is an important material in cooking and industrial oils.
The inedible parts of the rice plant can also be used: for example, the husks can be used as fertiliser or animal feed, and the straw for packaging or rug-making. In some areas, farmers retain rice stubble on their fields after harvesting so that it can be grazed by cattle. The utilisation of the harvests and the residues of rice were well-documented in the OECD revised consensus document on compositional considerations for new varieties of rice (Oryza sativa) (OECD, 2016).
Rice adapts to a wide range of weather and soil environments. The Asian cultivated rice, O. sativa, is distributed from latitude 53°N – beside the Amur River on the border between the Russian Federation (hereafter ‘Russia’) and China – to latitude 40°S, in central Argentina (IRRI, 1985). The two sub-species of O. sativa, indica and japonica, are cultivated in the plains of a tropical zone and a mid-latitude high-rainfall temperate zone respectively. Typical characteristics of the two sub-species are compared in Table 4.2. (Watanabe, 1997). In the regions where seasonal flooding occurs, such as the deltas of Bangladesh, East India, Thailand and Viet Nam, a part of indica rice may be grown as floating or deepwater rice (Catling, 1992).
Table 4.2. Comparison of the main characteristics of japonica and indica rice
|
Character |
japonica rice |
indica rice |
|
|---|---|---|---|
|
1 |
Leaf shape and colour |
Narrow and dark green |
Wide and light green |
|
2 |
Angle of flag leaf and rachis |
Large |
Small |
|
3 |
Culm length |
Short |
Long |
|
4 |
Culm strength |
Lithe and hard to break |
Hard and easy to break |
|
5 |
Lodging property of culm |
Hard to lodge |
Easy to lodge |
|
6 |
Grain shape |
Wide and thick and round cross-section |
Long, narrow and slightly flat |
|
7 |
Shattering habit |
Low shattering |
High shattering |
|
8 |
Awns |
Mostly awnless, a few varieties with short awns |
Awned with a variation of length |
|
9 |
Length and number of glume trichomes |
Relatively dense and short |
Not dense and relatively long |
|
10 |
Lengthwise ratio of grain |
2.5 or less |
2.5 or more |
|
11 |
Germination |
Slow |
Quick |
|
12 |
Phenol reactions |
- |
+ |
|
13 |
Potassium chlorate resistance |
High |
Susceptible |
|
14 |
Low-temperature tolerance |
High |
Susceptible |
|
15 |
Drought resistance |
Low |
High |
|
16 |
Endosperm destruction by alkali |
Easy |
Hard |
Source: Adapted from Matsuo, T. (1952), “Genecological studies on cultivated rice”, Bulletin of the National Institute of Agricultural Sciences Series D3, pp. 1-111 (in Japanese).
The highest latitude at which rice is cultivated is in Heilongjiang Province, China and they grow early flowering cultivars with no photoperiod sensitivity. Sufficient growth volume is secured in the short summers as the long day length provides enough sunlight and the inland climate has high enough temperatures.
In contrast, in low-latitude areas, there are two types of cultivars. One type is strongly photosensitive and floral transition is initiated by sensing the short day length that coincides with the end of the rainy season. The other is the improved mid-flowering type, which has lost its photosensitivity and grows even in winter, with double or triple cropping thanks to an abundance of sunlight. On the other hand, the African species, O. glaberrima, is distributed in the basin of the Niger River in Sub-Saharan Africa, where it has adapted to high temperatures and a dry environment.
As paddy fields are excellent at maintaining high circulation levels for water and nutrients, the growth of lowland rice is not influenced as much by soil properties as the growth of dryland crops. Although rice can survive in saline, alkaline and acidic soils containing sulphur compounds, it prefers semi-acidic (pH 6.0‑6.5) alluvial soils with a high degree of water retention.
In terms of water supplementation methods, there are four cropping systems: irrigated lowland (54% of the total world rice cultivation), rainfed lowland (25%), rainfed upland (13%) and deep water (8%). There are two systems of seedling establishment – direct seeding and transplanting – and system selection depends on the flatness of the field, the presence of irrigation systems, access to transplanting machines and the characteristics of the available cultivars. The use of direct seeding with small aircraft is widespread in the central plains of Australia and the United States.
Brown rice grains consist of bran layers (including the pericarp), an embryo and an endosperm. The endosperm consists of an aleurone layer and starch storage cells. Generally, starch in non-glutinous rice contains 10-30% amylose and 70-90% amylopectin. In glutinous rice, starch is composed of less than 5% amylose and mostly of amylopectin (Juliano and Villareal, 1993). The protein percentage ranges from 5% to 17% on a dry matter basis and the major protein in rice is glutelin (Juliano et al., 1968; Juliano, 1985). Fat, cellulose, minerals and vitamins are also present in brown rice (OECD, 2016).
As rice has a higher percentage of edible parts and a higher energy-conversion efficiency compared to other plants and because it is easily stored, it is a major source of calories in developing countries. Rice feeds more than half of the world’s population and accounts for 20% of the total energy needs of humans, compared with 19% for wheat and 5% for maize in calorie consumption (FAO, 2005). Moreover, a diverse food culture has developed owing to the various methods of cooking with glutinous and non-glutinous rice.
Statistical data on the global cultivation areas and production are listed in Table 4.3. Total production in 2020 was 758.49 million tonnes and the largest producer was China, followed in decreasing order by India, Bangladesh, Indonesia and Viet Nam (FAOSTAT, 2020). The total global area under cultivation was 164.45 million hectares, with the greatest area in India, followed by China, Bangladesh, Indonesia and Thailand. The average yield was estimated 4.6 t/ha for that year, but this varied widely depending on the sunlight, soil conditions, rice cultivar and cultivation system. High-yielding areas commonly have a large supply of nutrients and water from upstream, as well as flat land with high levels of sunlight. The highest-yielding country is Australia (10 t/ha), followed in order by Tajikistan, Egypt, Uruguay and the United States. Rice is a subsistence crop in most countries, whereas other cereals, such as wheat, soybean and corn, are mainly supplied as commercial crops. According to the FAOSTAT (2020), typical exporting countries are India, Viet Nam and Thailand, and typical importing countries are China, Philippines and Saudi Arabia, although these roles change frequently with alterations in the balance between domestic production and consumption.
Table 4.3. Production and cultivation of rice in the world, 2020
|
Country |
Production (1 000 t) |
Country |
Area (1 000 ha) |
||
|---|---|---|---|---|---|
|
1 |
China |
213 611 |
1 |
India |
45 000 |
|
2 |
India |
178 305 |
2 |
China |
30 342 |
|
3 |
Bangladesh |
54 906 |
3 |
Bangladesh |
11 418 |
|
4 |
Indonesia |
54 649 |
4 |
Indonesia |
10 657 |
|
5 |
Viet Nam |
42 759 |
5 |
Thailand |
10 402 |
|
6 |
Thailand |
30 231 |
6 |
Viet Nam |
7 223 |
|
7 |
Myanmar |
25 100 |
7 |
Myanmar |
6 656 |
|
8 |
Philippines |
19 295 |
8 |
Nigeria |
5 257 |
|
9 |
Brazil |
11 091 |
9 |
Philippines |
4 719 |
|
10 |
Cambodia |
10 960 |
10 |
Pakistan |
3 335 |
Source: FAOSTAT (2020). “Crops”, https://www.fao.org/faostat/ (accessed 11 March 2022).
Centres of origin, geographical distribution and agronomic practices
Centres of origin and diversity, and geographical distribution
Asian cultivated rice landraces have a high level of genetic diversity, and areas from Assam, India, to Yunnan Province, China, are centres of rice biodiversity (Oka, 1988c). Some cultivars can be cultivated in the Equatorial region, while others can only be cultivated in subarctic zones such as Heilongjiang Province in China and Hokkaido in Japan, which indicates that each cultivar or strain has distinct locations for suitable cultivation. Based on the high-throughput sequencing data of more than 3 000 cultivars (3 000 genomes), Asian cultivated rice (O. sativa) can be divided into several sub-species and ecotypes containing japonica, indica, Aus and Basmati (Huang et al., 2012; Wang et al., 2018). Among them, the genetic distance timing between japonica and indica sub-species is estimated to be more than 350 000 years, suggesting complex domestication processes in rice (Figure 4.5).
The genetic diversity among rice cultivars that has formed after rice domestication is believed to have started 10 000-20 000 years ago and was extensively investigated using the 3 000 genome information (Wang et al., 2018). Bioinformatics analyses of genome sequences for more than 400 accessions of ancestral species of Asian cultivated rice (O. rufipogon) have revealed that the genomes of the indica accessions are very similar to some O. rufipogon accessions (these are same as O. nivara in Figure 4.5; O. nivara is also known as annual O. rufipogon), while the genomes of tropical and temperate japonica have clear evolutionary distances from all tested O. rufipogon lines (Huang et al., 2012). It is assumed that
natural crossings between ancestral species of indica rice and O. rufipogon were caused presumptive gene flows.
Figure 4.5. Establishment of Asian cultivated rice and some key introgression events
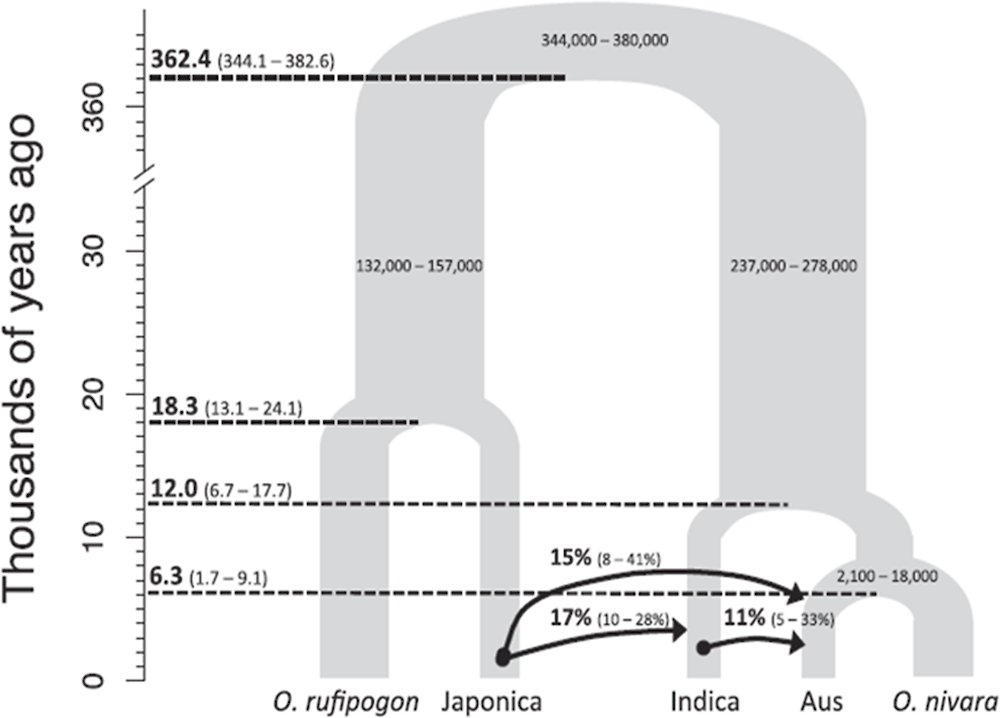
Note: Parameters were determined by a coalescent model. Left numbers are estimations of branching time. Arrows indicate the directions of introgressions. The numbers with the arrows are a median of estimation for introgressed genomic regions. The numbers in parenthesis indicate 95% confidence levels.
Source: Choi, J.Y. et al. (2017), “The rice paradox: Multiple origins but single domestication in Asian rice”, https://doi.org/10.1093/molbev/msx049.
In addition, key natural variations in the genes selected for by humans during rice domestication have been identified, including sh4 (seed shattering) (Li, Zhou and Sang, 2006), PROG1 (erect leaf stature) (Jin et al., 2008; Tan et al., 2008), rc (rice pericarp colour) (Sweeney et al., 2007), wx (stickiness of cooked rice) (Isshiki et al., 1998), qSH1 (seed shattering) (Konishi et al., 2006), Kala4 (black pericarp colour) (Oikawa et al., 2015) and LABA1 (seed awn) (Hua et al., 2015), via quantitative trait locus (QTL) analyses and subsequent fine mapping. In some cases, there are clear remains of critical introgression events of genomic fragments having natural variations from an ancient sub-species. Although there are several controversial models proposed, it is strongly supported that japonica sub-species have been domesticated first and then key introgression events may have occurred through natural backcrossing to establish other sub-species/ecotypes, such as indica and Aus (Figure 4.5) (Choi et al., 2017; Chen et al., 2019)
The lower Yangtze River valley is a likely candidate area for the origin of rice domestication. This is supported by the discovery of a large-scale paddy field cultivation of rice around 8 000 years ago and this is mainly based on archaeological analysis of the remains of old paddy fields (Fuller et al., 2009). Furthermore, by analysing vascular bundles in rice antiquity grains found in historic remains, the natural variations in the qSH1 seed shattering gene were found to have been selected in the lower valley area of the Yangtze River to reduce seed shattering (Zheng et al., 2016). On the other hand, as some tropical japonica cultivars have not been selected for key domestication-related genes, cultivation areas for the old tropical japonica cultivars may also be candidate areas for rice domestication (Konishi, Ebana and Izawa, 2008). The lower valley of Pearl River is also a candidate centre for rice domestication as some wild rice accessions of O. rufipogon, with genome sequences of 55 domestication sweep regions that are the most similar to those of some japonica cultivars, still grow wildly near the valley of Pearl River in the southern region of China (Huang et al., 2012). While all of this information is highly valuable, more research is required for a comprehensive understanding of the rice domestication processes.
African cultivated rice O. glaberrima, however, is thought to be derived from O. barthii, which is a wild rice species growing mainly in West African regions (Wang et al., 2014). It is believed that O. glaberrima has been domesticated in the delta region of the Niger River but rich genetic diversity has been observed in the upper swampy area of the Niger River. During the domestication of African cultivated rice, a few independent events that caused partial loss of seed shattering traits might have occurred, suggesting a complex domestication process (Choi et al., 2019). In East Africa, admix cultivars between O. sativa and O. glaberrima, termed New Rice for Africa (NERICA), have become promising owing to their higher productivity levels when compared with the local O. glaberrima landraces; however, critical improvements of the NERICA cultivars are still required (Yamamoto et al., 2018).
Weedy rice is geographically distributed in almost all rice-growing areas, such as in Brazil, Cambodia, China, Hungary, India, Italy, Japan, Korea, the Lao People’s Democratic Republic, Malaysia, the Philippines, Sri Lanka, Thailand, the United States and Viet Nam, under different cultivation systems including upland/lowland, transplanted/wet sown/dry sown, and so forth (Kraehmer et al., 2016).
Ecosystems and habitats where the species occurs natively, and where it has naturalised
Asian cultivated rice, O. sativa, is derived from Asian wild rice O. rufipogon (Oka, 1988a). More than 100 000 entries for local varieties and breeding lines have been maintained in the worldwide gene banks (IRRI.org Archived, 2012). These cultivars share similar characteristics with wild rice, except for their domestication-related traits, such as seed shattering, seed dormancy and plant architecture. As wild rice has survived by adjusting to tropical environments, rice cultivars can also grow year-round under tropical conditions. If rice experiences low temperatures at the panicle initiation stage, seed fertility will decrease because of aberrant meiosis in the pollen mother cells (Nishiyama, 1995). Therefore, in low temperate and high-latitude temperate areas, some rice cultivars can be grown only in summer with high temperatures.
Rice is a short-day plant that produces flowers when the daylight period becomes short. In low-latitude tropical areas and high-latitude temperate areas, critical day-lengths of the photosensitive cultivars are around 13-13.5 and 14-14.5 hours respectively (Oka, 1954). This indicates that cultivars in low latitudes tend to have shorter critical day-lengths. Therefore, temperate cultivars planted in tropical areas often generate fewer tillers and panicles and most tropical cultivars cannot generate ear panicles under temperate conditions. After domestication, cultivated rice was distributed to various areas and varieties with local adaptations have been selected for. Such local varieties can be transferred to places with similar environmental conditions.
Modern breeding varieties with many useful traits have been produced through crossbreeding. They can be introduced to both tropical and temperate areas if they are grown under appropriate cultivation conditions for day length and temperature.
Asian cultivated rice, O. sativa, is mainly divided into indica and japonica sub-species. Varieties of indica are predominantly cultivated in the delta and plain regions in Southeast Asia and South Asia. While many japonica varieties are grown in East Asia. Upland varieties classified as tropical japonica are observed in the mountain areas in Indochina and tropical islands. In Africa, most of the paddy rice and rainfed rice is indica varieties, whereas tropical japonica is dominant among the upland varieties. African cultivated rice, O. glaberrima, is also planted but its cultivation area is more restricted to the west-coast area including Guinea, Senegal and Sierra Leone, than that of O. sativa (Linares, 2002).
Agronomic and other intensively managed ecosystems where the species is grown or occurs on its own, including management practices
Rice can grow in a wide range of hydrological and climate conditions, ranging from flooded to water deficit conditions. The growing environments of cultivated rice are categorised into irrigated lowland, rainfed lowland, rainfed upland and deep water. More than 75% of the global rice grain yield is produced in irrigated lowlands that make up 54% of its cultivation area (GRiSP, 2013; Saito et al., 2013). After irrigated lowlands, rainfed lowlands produce 20% of the global rice grain yield. About 90% of the production comes from Asia and 4-5% from Africa and Latin America. China is the largest producer, followed by India. Major growing environments differ between regions and countries worldwide. Irrigated lowland areas are dominant in Asia and America, whereas rainfed lowland and upland areas are major growing environments in Africa. Deepwater rice grows in the Deltas of the Brahmaputra, Ganga and Mekong where the water depth is greater than 50 cm. Floating rice grows in flooded areas in which the water depth exceeds 100 cm and remains at these depths for several months. Rice also grows in mangrove swamps in some parts of West Africa, particularly in tidal estuaries close to the sea.
Growing environments and management practices affect the growth and yield ability of rice plants. In general, irrigated lowland rice produces higher yields than other growing environments, because high-yielding semi-dwarf cultivars are adopted, there are high inputs of chemical fertilisers and irrigation contributes to the reduction of drought and flooding risks. Lower rice yields in growing environments other than irrigated lowlands are caused by the high risk of drought or flooding. Farmers tend to apply smaller amounts of fertiliser and other inputs in case of these conditions (Saito et al., 2018).
In most rainfed upland areas, the yield is less than 2 t/ha. In contrast, the highest rice yield is 9.8 t/ha in irrigated lowlands in high-latitude areas where rice is grown during long summer days, such as California (US) and Southwest Australia and, at low latitude, areas having high diurnal temperatures and strong sunlight, such as Yunnan Province in China, the Nile Delta in Egypt and Uruguay. In these areas, rice is cultivated once per year.
In the tropics, rice can be cultivated throughout the year, with two or three crops when irrigation water is available. In places where climate conditions clearly differ between the dry and wet seasons, the rice yield is higher in the dry season because of the increased sunlight.
In Asia, rice is cultivated in the vast delta regions in Bangladesh, Cambodia, East India, Myanmar, Thailand and Viet Nam. Although these regions have abundant water resources for rice production, they did not benefit from the Green Revolution because of flooding problems and water control difficulties. However, since low-cost pump technology and short-duration rice cultivars were introduced, rice production systems in the delta regions have shifted from low-production deepwater rice and floating rice to high-production irrigated lowland rice (GRiSP, 2013). However, the damage caused by flooding and salinity owing to rising sea levels remains a serious concern in these regions. In regions where water resources are limited, such as China and north-western parts of the Hindustan Plain, the sustainable use of water resources is required.
Rainfed lowland rice mainly grows in South and Southeast Asia and Africa. Its cultivation is strongly affected by rainfall in the wet season from May to November in the northern hemisphere. Rainfed lowland rice is cropped in coastal areas, inland valleys and delta regions in the above-mentioned areas.
Rainfed upland rice grows in Africa, Asia and Latin America. Among them, Asia and Africa occupy 65% and 25% of the total rainfed upland rice area respectively (Saito et al., 2018). Rainfed upland rice occupies 32% of the total rice area in Africa but only 6% in Asia. During the past 30 years, the upland rice area has increased in Africa, while it has reduced in Asia and Latin America.
Some resource-poor countries with higher annual rainfall and lower gross national incomes per capita tend to have a higher percentage of upland rice in their total area of rice cultivation. In Asia, upland rice cultivation areas are high in India, Indonesia, and China.
Upland rice was historically cultivated in shifting cultivation patterns on hilly slopes in the mountainous region of the Indochina Peninsula. In this region, high population pressure has caused the change from shifting cultivation to permanent farming systems in the limited fields.
In tropical Asia, cattle and water buffalo have been used for land preparation for many years. However, the use of agricultural machines such as hand tractors has become more popular with rice farmers. There are two crop establishment methods: direct seeding and transplanting.
Direct seeding dominates in South Asia, including Bangladesh, India and Sri Lanka. In Southeast Asia, transplanting is a major crop establishment method in inland areas, while direct seeding dominates in delta areas. In Africa, transplanting is widely used in irrigated lowlands, while direct seeding is common in rainfed conditions (Niang et al., 2017). Several different methods of direct seeding, such as broadcasting, dibbling and drilling, are used in the rainfed upland areas of Africa and Asia. Direct seeding areas have increased in both the irrigated and rainfed lowlands in Asia because of labour shortages. Direct seeding is a common crop establishment method in deepwater rice.
Transplanting is also used to avoid damage from flooding and weed infestation in some areas (Pandey and Velasco, 2005). For large-scale rice farming in the United States and Latin America, rice is directly sown in wet or dry fields using aircrafts or large seeding machines. Double rice cropping is practised by using ratooning in the southern states of the United States. In tropical Africa and Asia, transplanting and direct seeding are practised by hand, as the use of agricultural machines is still limited, while agricultural chemicals, such as fertiliser and herbicides, are commonly used (Rodenburg et al., 2019).
In the tropical areas of Asia and Africa, the use of agricultural machines for harvesting is still limited, except in Thailand, Viet Nam, and Senegal. Rice is thus usually harvested by hand. Harvested rice grains are usually threshed in the field using threshing equipment with or without a power source. In some areas, harvested panicle bundles are taken from the field for storage without threshing.
Large genetic variations among rice cultivars are observed in seed dormancy after ripening. Seed dormancy affects seed longevity and spontaneous growth as strong dormancy makes it possible to germinate and survive by avoiding unfavourable growth periods.
There is broad genetic variation at the level of seed shattering. In Japan, non-shattering cultivars have been selected to avoid grain loss in the field owing to the combine harvester. On the other hand, easy shattering cultivars are widely cultivated in tropical Asia, where some seeds drop on the ground during harvesting and threshing in the field. They germinate in the next cropping season and result in an increased risk of contamination of different rice cultivars and spontaneous growth.
Where cultivated rice is grown near populations of wild rice, hybrid populations derived from outcrossing between cultivated and wild rice are sometimes observed. Individual plants from hybrid populations have broad ranges of phenotypic variation in plant height, hull colour, awn length, seed colour and shattering habit. Some weedy rice lines have also originated from outcrossing between cultivated and wild rice (Akasaka et al., 2009; Brozynska et al., 2017).
Weedy rice includes wild rice, off-types of cultivated rice and hybrid swarm as volunteers from seeds that shattered in the previous cropping and germinated during the following rice-growing season. They frequently continue to grow repeatedly via fallen seeds. It is often observed in paddy fields. As the weedy rice has similar morphological and physiological characteristics to the rice cultivars, it is difficult to avoid contamination of crops during harvesting periods, causing yield loss, caused by competition during the growth stage and seed shattering, and the decline of grain quality. The management of weedy rice is more difficult with direct seeding methods than transplanting systems. It is an emerging problem in Asia as the direct seeding method has become more widespread than transplanting systems (Chauhan, 2013).
To control weedy rice, integrated weed management measures including the rotation of direct seeding and transplanting, rotation with other crops, the use of herbicides, puddling paddy fields before transplanting and manually pulling out weedy rice, are required. In addition, a wide range of herbicide susceptibility exists in cultivated rice, e.g. indica vs. japonica to acetolactate synthase (ALS) inhibitors (Kobayashi, Yogo and Sugiyama, 1995), and whether having functional his1 gene which confers resistance to 4‑HPPD inhibitors (Maeda et al., 2019). Moreover, herbicide-resistant rice cultivars are more frequently used as one of the management options in the United States and Latin America (Sudianto et al., 2013).
Reproductive biology
Generation time and duration under natural circumstances, and where grown or managed
Generation times for cultivated rice differ greatly among the varieties, ranging from approximately three to six months. During the vegetative growth period, which is followed by the reproductive growth period, the plant develops a dozen leaves and tillers. Environmental conditions, such as day length and temperature, affect plant growth and phase transitions to reproductive growth. Generally, long-day conditions lower plant height and deepen the green colour of the leaves. By contrast, short-day conditions are often required for phase transitions to reproductive growth.
Rice is commonly cultivated once per year and self-fertilised seeds are harvested. However, there are many O. sativa varieties that can be maintained as vegetative clones using axillary buds after bearing seeds, depending on the conditions. This characteristic enables newly elongated tillers from harvested stocks to grow again. Such tillers, called ratoons, can be harvested again. It is thought that this perennial property is derived from O. rufipogon, an ancestor species of Asian cultivated rice (O. sativa) (Morishima, Hinata and Oka, 1963). By contrast, another cultivated species (O. glaberrima) possesses more annual properties (Morishima, Hinata and Oka, 1962).
Reproduction (production of flowers, seeds and vegetative propagules)
Reproductive structure
O. sativa is a self-pollinating plant. A single rice flower, called a spikelet, contains 6 anthers, harbouring more than 1 000 pollen grains, and a pistil with furcate styles, each leading to stigmas. In concurrence with the opening of rice spikelets, pollen grains fall onto the stigmas, germinate and elongate their pollen tubes. One of the pollen tubes that reach the embryo sac takes part in double fertilisation.
The spikelet opening of rice starts on the day of or the day following panicle emergence. Spikelet opening proceeds from the panicle tip to the basal part (Moldenhauer and Gibbons, 2003). In detail, the top primary rachis-branch starts to open first and then the lower primary rachis-branches start to open in sequential order. In a primary rachis-branch, the top spikelet opens first and the lowest spikelet in the same primary rachis-branch opens next. Then, the lower spikelets start to open in sequence and the second-highest spikelet opens last. The spikelets in the secondary rachis-branches also start to open from the top and the sequence of spikelet opening in the secondary rachis-branch is the same as the one in the primary branch. The sequence of spikelet opening is identical to that of the differentiation of the flower in the young panicle (Hoshikawa, 1989).
Yoshida (1981) reported that it takes five days for most spikelets in a single panicle to open and seven to ten days for all spikelets. Sleper and Poehlman (2006) reported that the spikelet opening period of a single panicle lasts from three to seven days after the heading and most of them bloom two to four days after the spikelet opening begins.
The spikelet opening process of cultivated rice is as follows: immediately before spikelet opening, the filaments of the stamens become elongated and the anthers move to the upper part of the spikelet. Simultaneously, the stigmas standing straight begin to open and also their branches extend outward to increase the area available to receive pollen grains. The lodicule at the bottom of the palea takes in water and swells, and the swelling pressure pushes out the lemma. At the same time, the interlocking between the palea and the lemma is undone, and the top edges of the spikelet gradually start to open. Then spikelet opening starts. Anthers that reach the top of the spikelet start to dehisce and the pollen grains fall onto the pistil of the same spikelet. In most cases, the pollination process is complete at this stage. After that, the anthers are put out of the spikelet through the further opening of the lemma and the palea, and continuous elongation of the stamen filaments.
Extruded anthers release resting pollen grains to the outside. In 10‑25 minutes after the spikelet opening starts, the opening between the lemma and the palea expands to an angle of 25‑30 degrees. When the spikelet fully opens, both stigmas spread to an angle of 90 degrees compared to the apical-basal axis of the pistil and their apices become exposed through the opening between the lemma and the palea (Hoshikawa, 1989).
After spikelet opening, loss of moisture and subsequent shrinking of the lodicule cause the lemma to return to its previous position, resulting in spikelet closure, which terminates the spikelet opening process. Anthers and filaments extruded out of the spikelet remain outside of the spikelet. The spikelet opening period of a single spikelet ranges between 1 and 2.5 hours (Hoshikawa, 1989).
Although there are individual differences, pollen grains that fall on the stigmas start to germinate after two to three minutes in the shortest case and pollen tubes become elongated to the ovule in the ovary through the style. Under suitable conditions, the tip of the pollen tube reaches inside of the embryo sac within 15 minutes and then the fertilisation processes are completed during the next five to six hours (Hoshikawa, 1989).
The time of spikelet opening for cultivated rice varies depending on the weather conditions and genetic characteristics. Rice spikelet opening normally occurs between 9 a.m. and 2 p.m. (Moldenhauer and Gibbons, 2003) or 10 a.m. and 2 p.m. (Sleper and Poehlman, 2006). The spikelet opening times in tropical areas tend to be longer than those observed in temperate areas (Nagai, 1959). In detail, Hoshikawa (1989) reported that in temperate areas rice spikelet opening starts around 9 a.m., the peak of spikelet opening is around 11 a.m. and most spikelets close around 1 p.m. when the weather is fine and the temperatures are high enough. However, when the temperatures are around 20°C, spikelet opening starts around noon, lasts sluggishly until around 5 p.m. and ends around 6 p.m.
Yoshida (1981) and Moldenhauer and Gibbons (2003) also reported that the beginning and the end of spikelet opening could be delayed by low temperatures and cloudy conditions. Additionally, in severe weather, rice spikelets do not open but the elongation of the filaments and dehiscence of the anthers take place inside the spikelet, resulting in pollination without spikelet opening, which is called cleistogamy (Hoshikawa, 1989).
Yoshida (1981) reported genetic differences for the start and end times of rice spikelet opening in tropical areas. While the spikelet opening of O. sativa starts around 8 a.m. and ends around 1 p.m., the spikelet of O. glaberrima starts to open earlier at around 7 a.m. and ends after a shorter duration, around 11 a.m. There are also wild rice varieties whose spikelets start to open in the early morning or at night (Watanabe, 1993).
Pollination, pollen dispersal, pollen viability
As described in the previous subsection on reproductive structure, rice is a self-pollinating plant. However, natural crossings of rice can occur by the wind. When pollen fertility of recipient plants is decreased by low-temperature conditions in the pollen formation period, the crossing rate rises (Sato and Yokoya, 2008; Tanno et al., 2011). The crossing rate of cultivated rice is affected by other conditions, including the duration of spikelet opening, wind direction and speed, and the scale of the pollen source.
It is thought that the differences in the morphological characteristics of the stamens and pistils are responsible for the differences in the natural crossing rates of the rice. In a study on the seed production of hybrid rice cultivars, Virmani (1994) reported that the crossing rate was increased when the anthers and stigmas were larger, and with a higher frequency of exposure of the stigma out of the spikelet, which increased the probability of catching pollen in the air. These characteristics are more common in wild rice than in cultivated rice (Oka and Morishima, 1967; Uga et al., 2003). The length of anthers is highly correlated with the number of enclosed pollen grains. A single anther of cultivated or wild rice can contain approximately 700-2 500 or 700-9 000 pollen grains respectively (Oka and Morishima, 1967). Bakti and Tanaka (2019) reported that O. rufipogon, a wild rice species, tends to expose its stigma outside of the spikelet in contrast to cultivated rice.
The morphology of the panicle and the positional relationship between the panicle and the flag leaf also affect the natural crossing rate (Virmani, 1994). Some wild rice varieties have a time lag between spikelet opening and the release of pollen grains. This also contributes to an increase in the natural crossing rate (Oka and Morishima, 1967).
Nagao and Takano (1938) and Oka and Morishima (1967) characterised the viability time for pollen grains. In cultivated rice, the rate of fertilisation drops over time after the release of pollen grains from the anther, as the released pollen grains become infertile after five minutes. An immediate decrease in the number of fertile pollen grains after their release from the anther was also observed on artificial growth medium during germination tests and vital staining. It is considered that the loss of pollen viability results from desiccation (Nakayama, 1934; Koga et al., 1971; Khatun and Flowers, 1995). By contrast, in wild rice, fertilisation can occur in less than nine minutes after pollen grain release (Oka and Morishima, 1967).
However, once the stigma becomes competent, it maintains competency for three to seven days (Yoshida, 1981). Therefore, the natural crossing rate increases if the stigma remains out of the spikelet after the end of the spikelet opening (Kato and Namai, 1987; Xu and Shen, 1987; Yan and Li, 1987; Yan et al., 2009). Nagao and Takano (1938) found that the fertilisation ability of stigmas is greatly decreased three days after spikelet opening in artificial crossing experiments.
Seed production and natural dispersal of seeds
Completion in 2004 of the genome sequencing project for the rice cultivar, Nipponbare, enabled extensive analyses of the quantitative trait loci (QTL) between cultivars and various mutants related to the morphology and development of inflorescence and seed formation in rice. This work allowed for the identification of many of the causal genes regulating the morphology of seeds and panicles in rice.
It is well known that the characteristics controlling the variations of both seed length and width are genetically regulated in an independent manner (Zuo and Li, 2014). The differences in seed shape observed in various rice cultivars are commonly regulated by such characteristic QTL. Among the genes regulating seed width, qSW5/GW5 is known to contribute markedly to rice variation and has been revealed to function as a genetic element in brassinosteroid signalling (Shomura et al., 2008; Weng et al., 2008; Liu et al., 2017a). Since qSW5/GW5 narrows seed width, it is believed that the loss of these functional alleles was selected for during the early stages of rice domestication. Another main regulator gene of seed length in rice is GS3, which encodes a G protein γ‑subunit (Fan et al., 2009; Mao et al., 2010).
Several genes related to awn formation have been identified. In the early stages of rice domestication, the defective alleles of An-1 were selected and, subsequently, the defective alleles of RAE2 and An-2 were selected (Luo et al., 2013; Bessho‑Uehara et al., 2016; Gu et al., 2015). This has led to the loss of awns in the spikelets of most rice cultivars.
Furthermore, Gn1a (Ashikari et al., 2005), WFP/IPA (Jiao et al., 2010; Miura et al., 2010) and APO1 (Ikeda et al., 2007) function to control the panicle size and seed number (or the number of spikelets in a panicle). Gn1a encodes an enzyme for a phytohormone, cytokinin, whereas WFP/IPA encodes an SPL (SQUAMOSA Promoter binding protein-like)-type transcriptional factor. The APO1 gene encodes an F‑box protein related to specific protein degradation and is assigned as an orthologue of the UFO gene in Arabidopsis thaliana. In addition, it is known that the WFP/IPA gene is regulated epigenetically and affected by specific miRNAs.
Since sexual reproduction in rice is mainly mediated by self-pollination, there is no significant agricultural problem unless the fertility of plants decreases due to specific environmental conditions and/or genetic effects. Rice plants with the spw1-cls allele have a mutation in the SPW1 (SUPERWOMAN1) gene that results in an amino acid change and consequently, a cleistogamous trait is exhibited (Yoshida et al., 2007). The use of this SPW1 allele has been considered for genetically engineered (GE) rice cultivation. However, there is low stability of the cleistogamy trait under relatively low-temperature conditions and, consequently, its economic uses have not been pursued.
In wild rice, the abscission layers are formed at the base of the spikelet. After pollination, the layers start to be degraded and maturing seeds shatter to propagate seeds in the natural environment. In cultivated rice, non-shattering traits have been selected by humans. There are several natural mutations in various distinct genes that are involved in the diversity of seed shattering traits in rice cultivars. There are broad variations of these traits among different species/sub-species/varieties. The easy shattering cultivars, including many of the indica cultivars, are grown in developing countries with less access to agricultural implements and machinery. Most japonica cultivars, however, exhibit non-shattering traits, making them suitable for use with agricultural machinery. During domestication, a defective allele in sh4, a standing variation in wild rice, has been strongly selected for. Consequently, all of the cultivars tested have the same sh4 allele (Li, Zhou and Sang, 2006).
In addition to the defective sh4 allele, the non-shattering trait in most of the currently used Japanese cultivars is due to the natural variations in the qSH1 gene (Konishi et al., 2006) and this mutation has been observed only in temperate japonica cultivars. Compared with the selection for the sh4 gene, this mutation in qSH1 was selected for during the establishment of the cultivar, or temperate japonica. Based on the analysis of the antique carbonised rice grains found in the paddy field remains from the lower valley of the Yangtze River, it is speculated that this mutation was selected approximately 7 500 years ago (Zheng et al., 2016). Rice plants having both defective alleles possess no abscission layer, making the cultivars suitable only for mechanical harvesting. Since qSH1 is normally expressed at the shoot apex region and functions in the development and maintenance of the shoot meristems in rice, the selected natural mutation resides in the cis-regulatory region of the qSH1 promoter and represses qSH1 transcription only at the provisional abscission layers (Konishi et al., 2006).
There have been two genes identified as causal genes involved in the loss of seed shattering during the domestication of African cultivated rice (O. glaberrima). One is an orthologue of the sh4 gene in Asian cultivated rice (O. sativa), the other is SH3, which encodes a YABBY-type transcription factor. Since the standing variations still exist in African wild rice (O. barthii), it is speculated that those mutations were selected sequentially to lose seed shattering traits in African cultivated rice (O. glaberrima) (Wu et al., 2017; Lv et al., 2018).
Seed viability, longevity, dormancy, natural seed bank, germination and seedling viability and establishment
The seeds of the varieties with strong dormancy maintain their viability for several seasons. For example, Surjamkhi, a cultivar with a strong dormancy, maintained viability after six years, whereas Fujiminori, a cultivar with weak dormancy, lost viability after three years (Takahashi and Suzuki, 1975). Seeds with no or weak dormancy germinate in the ear of standing rice (vivipary). When vivipary occurs, the grains lose their value as food. In the past, farmers and breeders have continuously selected cultivars with appropriate dormancy for their cultivation styles (Bewley et al., 2013).
The seeds of the varieties with strong dormancy can become weedy because the volunteer seeds from previous seasons would germinate sporadically in the field where new cultivars are grown. The factors responsible for seed dormancy reside in the chaff. The dormancy of seeds increases the potential weediness of the cultivars when the seeds are released from the ear. Indica cultivars have a wider range of dormancy than japonica cultivars and often become indigenous weeds. Red-kernelled rice cultivars also shed easily and have strong dormancy in both indica and japonica. They consequently become weeds frequently in rice fields, creating problems for farmers. The intercrossing between cultivated rice and indigenous weeds, including red-kernelled rice, occurs in many rice farming countries and has also become a widespread problem (Ziska et al., 2015).
The optimal temperature for rice seed germination ranges from 13°C to 30°C, and the highest temperature is 44°C; there are varietal differences associated with these variations. Varieties with outstanding germination properties at low temperatures are selected from wild varieties found in high-latitude regions. At optimal temperatures, rice seeds absorb approximately 25% of their air-dried seed weight in water and germinate in the presence of oxygen. Unlike other Poaceae (Gramineae) crops, rice seeds can germinate under conditions with low oxygen concentrations, through anaerobic respiration. It was thought that the germination of rice seeds was not affected by light. However, it has been reported that light promotes seed germination for some varieties of weedy rice (light-induced germination) (Chung and Paek, 2003). Although light weakly affects the promotion of seed germination in cultivated rice, there are varietal differences in the extent of the light induction (Lee et al., 2010).
The biological aspects of rice seeds in natural conditions have mostly been studied in weedy rice and volunteer rice. A difference was found in the survival rate of the seeds between the surface of the field and the soil layer. The survival rate of the seeds on the field surface dropped below 50% after 1 winter and all seeds died after 2 winters (Hosoi et al, 2010). The seeds buried in the soil layer (10-15 cm below ground), however, maintained their germination rate after 2 winters but died after 3 winters. It is known that the moisture content of the soil affects the maintenance of viability in buried conditions, with a report showing that the viability of red-kernelled rice seeds buried in irrigated fields was longer than when they were buried in non-irrigated fields (Suzuki, 2003).
In wild rice, the germination rate of seeds buried at 25°C for 40 months was higher than 40%, with the water content of the seeds exceeding 30%. However, when seeds were maintained at a water content of 16%, all seeds died after 16 months (Oka, 1992). In cultivated rice, it is known that viability decreases quickly in high-temperature and high-humidity conditions (Roberts, 1961). There are varietal differences in the viability of seeds. There are many reports in which seeds of indica rice cultivated in tropical areas tend to have a longer life duration than japonica rice cultivated in temperate areas at relatively high altitudes (Juliano, Perez and Chang, 1990; Chang, 1991; Ellis, Hong and Roberts, 1992; Rao and Jackson, 1996a, 1996b, 1996c, 1997; Ebina, Nakamura and Yamamoto, 1998; Padma and Reddy, 2000).
The sowing depth, that is the soil depth at which the seed starts to germinate, affects the germination of rice seeds (Ohno et al., 2018). In some cultivars, sowing depths that exceed five centimetres significantly inhibit germination. On the other hand, some weedy rice varieties can germinate even at 13 cm below ground, indicating that the tolerance of greater sowing depths is an effective trait for survival under natural conditions (Vidotto and Ferrero, 2000). Rice seeds can also germinate when submerged. However, when deeply submerged (2-8 cm), germination can be suppressed (Chauhan, 2012). In cultivated rice, red‑kernelled rice is superior in terms of its resistance to submerged conditions and is used as a genetic resource to improve the submergence tolerance at the germination stage of cultivated rice (Septiningsih et al., 2013).
Asexual propagation (apomixis, vegetative reproduction)
O. sativa is cultivated as an annual crop. However, the plants can continue their vegetative growth cycle after bearing if the water and temperature conditions are suitable. It is thought that the perennial property of O. sativa is derived from an ancestral species O. rufipogon (Morishima, Hinata and Oka, 1963). In natural conditions, the tiller buds at the basal nodes begin to elongate after the harvesting of the ears. The new tillering buds, called ratoons, can elongate in long-day conditions. In some countries such as Brazil, China, the Dominican Republic, India and the United StatesUS, farmers grow the ratoons and harvest a second crop of the grains.
The rhizome is another characteristic related to perennial rice. A wild rice species, O. longistaminata, is perennial and has a strong rhizomatous nature, and several loci were identified that controlled this trait (Hu et al., 2003; Zhang et al., 2015). Rice rhizomes have several characteristics, including that the buds bend to elongate horizontally and, as the rhizome expands, it maintains its juvenile phase (Yoshida et al., 2016).
The perennial properties of rice can be beneficial, including its rhizomatous nature which competes with weeds and is useful for improving the soil environment in non-ploughing cultures. Consequently, there is an ongoing effort to introduce these perennial properties into cultivated rice varieties (Sacks, 2013).
Although there are many Poaceae (Gramineae) species that reproduce by apomixis, no apomictic species have been identified in the genus Oryza (Khush et al., 1994). The productivity of rice can be improved by breeding and especially through first-generation hybrid breeding if the apomictic property was introduced into existing cultivars. Thus, the idea that the apomictic property could be introduced into rice from genetic resources was proposed. Recently, a technique has been developed that introduces apomictic reproduction into rice by genetically engineering the reproductive process through genome editing (Xie et al., 2019). The apomixis was introduced into rice by mutating four responsible genes for meiotic recombination and the quadruple mutant line was named Apomictic Offspring Producer (AOP).
It is possible to induce and propagate rice calluses using tissue and cell culture methods. In the appropriate conditions, calluses re-differentiate into tissues and plantlets, and propagate asexually. Haploid plants of rice can be easily obtained by pollen cultures. The haploid plants sometimes become diploid plants by natural duplication. Diploid plants can also be easily obtained by chemically treating haploid plants (Niizeki and Oono, 1968).
Genetics
Relevant detailed genetic information on the species
Gene pool
The Asian cultivated rice, O. sativa, is an AA genome diploid species (2n = 2x = 24). The primary and secondary gene pools of this species are defined based on their level of reproductive isolation (Khush, 1997; Jena, 2010). One cultivated species (O. glaberrima) and six wild species (O. rufipogon, O. nivara, O. longistaminata, O. barthii, O. glumaepatula, and O. meridionalis) constitute the primary gene pool. They share the AA genome and are crossable with O. sativa. These species with AA genomes correspond to those in the sativa complex defined by Morishima and Oka (1960), based on the morphological characteristics of the genus Oryza. Among them, O. rufipogon have high crossability with O. sativa, because they are wild progenitors of common cultivated rice. They grow mainly in swampy and wet areas in tropical Asia and gene flow is often observed between cultivated and wild rice around the paddy fields (Oka, 1988b). The African cultivated rice, O. glaberrima, can be crossed with O. sativa; however, their hybrids cannot produce fertile seeds due to severe pollen sterility (Sano, Chu and Oka, 1979; Garavito et al., 2010).
The secondary gene pool consists of wild species in the officinalis complex (Table 4.1). This complex includes 11 wild species having BB, CC, BBCC, CCDD and EE genomes, such as O. officinalis and O. minuta (Khush, 1997; Jena, 2010). Crosses between O. sativa and these species can be accomplished by embryo rescue using tissue culture techniques (Brar and Khush, 1997). Their hybrids are completely sterile because normal pairing between their chromosomes for the different genomes in meiosis cannot occur.
Genome information
The nuclear genomes of the O. sativa japonica cultivar Nipponbare and the indica cultivar 93-11 have been sequenced and assembled as reference genomes (IRGSP, 2005; Yu et al., 2002, 2005). The genome size of Nipponbare was estimated to be 389 Mb and that of 93-11 was 466 Mb. The reference genome of Nipponbare has been improved by adding sequence information derived from short-read high-throughput sequencing, resulting in the correction of sequence errors and increasing genome coverage (Kawahara et al., 2013). Genome databases of Nipponbare with detailed gene annotation information have been developed as RAP-DB (Sakai et al., 2013) and MSU (Ouyang et al., 2007). Subsequently, the genomes of the Japanese elite cultivar Koshihikari and the African cultivated species O. glaberrima were sequenced and assembled, using the Nipponbare and 93-11 as reference genomes (Yamamoto et al., 2010; Sakai et al., 2011). More than 3 000 diverse Asian accessions have been made to the gene bank of the International Rice Research Institute (IRRI), revealing a large amount of structural variation among them (Wang et al., 2018; Fuentes et al., 2019).
Based on de novo assembly from high-throughput sequencing data, relatively high-quality reference genomes have been assembled for Shuhui498 (Du et al., 2017), O. laberrima (Wang et al., 2014) and several wild relatives with AA genomes: O. rufipogon (Zhao et al., 2018), O. nivara, O. barthii, O. glumaepatula, and O. meridionalis (Zhang et al., 2014). In addition, The International Oryza Map Alignment Project (Jacquemin et al., 2013) has made available the sequences of wild species other than those with AA genomes, such as O. longistaminata (Reuscher et al., 2018), O. brachyantha (Chen et al., 2013) and O. granulata (Wu et al., 2018).
Expression profiling has also been conducted for rice and several databases are available. For the japonica cultivar Nipponbare, transcriptome data for different growth stages and tissues are available in RiceXPro (Sato et al., 2010, 2013) and its co-expression database RiceFREND (Sato et al., 2012). For indica cultivars, similar expression profiling databases are available on Zhenshan 97 and Minghui 63, the parental lines of the primary F1 hybrid variety grown in China (Wang et al., 2010). Proteome and metabolome databases were also constructed (Hong et al., 2019). As for the genetic DNA markers, 2 240 simple sequence repeat (SSR) markers have been identified and summarised (McCouch et al., 2002) and are widely used in genetic and molecular analyses and marker-assisted selection (MAS) in rice breeding. The resequencing of many cultivars identified single nucleotide polymorphisms (SNPs). These SNPs are used in genetic analysis and selection in breeding programmes (Huang et al., 2009, 2010; Eishire et al., 2011).
Genetic factors affecting maturity (heading date)
Heading date, or timing of heading, is the event during which the panicle emerges from the sheath of the final mature leaf, termed the flag leaf. The heading date can be considered an indicator of flowering time in rice and an important agricultural trait related to yield and suitability to cultivate in diverse geographical locations. Genetically, it is well known as a quantitative trait that is regulated by multiple loci. Based on genetic linkage analyses with known genetic markers in rice and using the progenies from between the cultivars, several loci having clear effects on heading date have been mapped using classical genetics approaches, such as Se1, E1, E2, E3 and Ef1, although this information has been considered fundamental knowledge and has not been used for breeding (Hori, Matsubara and Yano, 2016).
In the late 1990s, many DNA markers were developed and subsequently, many QTL analyses were performed. As a result, many of the QTL that control heading dates were identified. Particularly, Yano’s group in Japan performed an extensive QTL analysis using F2 progenies and backcrossed progenies between Nipponbare (a temperate japonica cultivar) and Kasalath (an Aus cultivar) and identified more than 15 QTL affecting heading date between them (Yano et al., 2001). Although a few of those QTL are speculated to be due to the natural variations in the genes previously identified, it is not easy to identify all the relationships between them. This is because the positions of the QTL are based on the positions of DNA markers, whereas the positions of the previously identified genetic loci related to heading date were defined with genetic distances based on other known genetic loci that are easily phenotyped, such as wx (waxy: glutinous endosperm), C (chromogen for anthocyanin), and Pl (purple leaf). At present, more than 14 heading time-related genes have been identified genetically using QTL cloning in rice (Hori, Matsubara and Yano, 2016) (Table 4.4). In recent years, using the precise positional information of identified QTL affecting heading date but not using any phenotypic data, DNA-marker-assisted breeding has been performed to develop new cultivars that have preferable heading dates for their given cultivation areas (Hori, Matsubara and Yano, 2016).
Among the many heading date QTL in rice that have already been identified, several that make large contributions to rice breeding due to their critical effects are discussed here. The Hd1 (Heading date 1) gene functions as a floral promoter under short-day conditions but as a floral repressor under long-day conditions (Yano et al., 2000). Thus, Hd1 is bifunctional and can contribute to local adaptions in temperate cultivation areas. For tropical japonica and the Aus ecotype that are cultivated in tropical and sub-tropical areas, a defective allele of Hd1 (hd1) has become dominant (Fujino et al., 2010). The hd1 allele causes prolonged vegetative growth and reduces the photoperiodic responses of the floral transitions in cultivation areas at low latitudes. This defect may help crops adjust to different seasons and avoid the flooding seasons for major cultivars in Aus ecotypes (Fujino et al., 2010).
Generally, rice breeding cultivation areas in Asia have historically progressed northward. In particular, a defective natural mutation occurred in the Ghd7 (Grain number and heading date 7) gene that contributed critically to the extension of rice cultivation into subarctic areas, such as Hokkaido in Japan and Heilongjiang Province in China (Xue et al., 2008). Ghd7 functions as a very strong floral repressor under long-day conditions. All tested cultivars adapted to the Hokkaido areas possessed the defective Ghd7 allele (ghd7). It has led to the development (or selection) of cultivars that flower in early August under long-day natural conditions with no responses to day length changes and consequently they are able to provide enough yield for the human populations in the subarctic climate of the Hokkaido area. Similarly, the defective alleles of the Dth8/Hd5 gene and OsPRR37/Hd2 gene have both clearly contributed to the northward progression of rice cultivation (Li et al., 2015). Both the reduction of photoperiod sensitivity and the early flowering phenotype due to the above natural variations may play pivotal roles in the progression of rice to the northern areas.
Table 4.4. Classical Mendelian genes and isolated genes for natural variation in heading date in rice
|
Gene symbol |
Synonym |
Effect on flowering1 |
Chromosome |
RAP ID2 |
MSU ID3 |
Description |
References4 |
|---|---|---|---|---|---|---|---|
|
Se |
Se1, K, Lm, Hd1 |
SD promotion/ LD repression |
6 |
Os06g0275000 |
LOC_Os06g16370 |
Zinc-finger protein |
Chandraratna (1953, 1955), Yokoo and Fujimaki (1971), Yano et al. (1997, 2000) |
|
E1 |
M, m-Ef1, Ghd7 |
LD repression |
7 |
Os07g0261200 |
LOC_Os07g15770 |
CCT (CONSTANS, CONSTANS-LIKE, and TIMING OF CHLOROPHYLL A/B BINDING1) domain protein |
Syakudo and Kawase (1953), Syakudo et al. (1954), Tsai and Oka (1966), Tsai (1976), Okumoto et al. (1992), Okumoto and Tanisaka (1997), Xue et al. (2008) |
|
E2 |
Hd17, Ef7, OsELF3-1, OsELF3, Hd-q |
SD/LD promotion |
6 |
Os06g0142600 |
LOC_Os06g05060 |
Homolog of Arabidopsis EARLY FLOWERING 3 protein |
Syakudo and Kawase (1953), Syakudo et al. (1954), Matsubara et al. (2008a), Monden et al. (2009), Yuan et al. (2009), Matsubara et al. (2012), Saito et al. (2012) |
|
E3 |
Hd6 |
LD repression |
3 |
Os03g0762000 |
LOC_Os03g55389 |
Similar to protein kinase CK2, alpha subunit |
Syakudo and Kawase (1953), Syakudo et al. (1954), Takahashi et al. (2001) |
|
E |
Ef1, Ehd1 |
SD/LD promotion |
10 |
Os10g0463400 |
LOC_Os10g32600 |
B-type response regulator |
Tsai and Oka (1966), Tsai (1976), Sato et al. (1988), Doi et al. (2004), Saito et al. (2009) |
|
Hd3a |
|
SD promotion |
6 |
Os06g0157700 |
LOC_Os06g06320 |
Florigen |
Kojima et al. (2002) |
|
RFT1 |
|
LD promotion |
6 |
Os06g0157500 |
LOC_Os06g06300 |
Florigen |
Kojima et al. (2002), Ogiso-Tanaka et al. (2013) |
|
DTH8 |
Ghd8, LHD1, Hd5, LH8 |
SD promotion/ LD repression |
8 |
Os08g0174500 |
LOC_Os08g07740 |
Putative HAP3 subunit of CCAAT box-binding transcription factor |
Wei et al. (2010), Dai et al. (2012), Fujino et al. (2013), Chen et al. (2014) |
|
DTH3 |
OsMADS50 |
SD/LD promotion |
3 |
Os03g0122600 |
LOC_Os03g03070; LOC_Os03g03100 |
MIKC-type MADS-box protein |
Lee et al. (2004), Bian et al. (2011) |
|
DTH2 |
|
LD promotion |
2 |
Os02g0724000 |
LOC_Os02g49230 |
CONSTANS-like protein |
Wu et al. (2013) |
|
Hd16 |
EL1 |
LD repression |
3 |
Os03g0793500 |
LOC_Os03g57940 |
Casein kinase I |
Dai and Xue (2010), Hori et al. (2013), Kwon et al. (2014) |
|
OsPRR37 |
Hd2 |
LD repression |
7 |
Os07g0695100 |
LOC_Os07g49460 |
Pseudo-response regulator |
Koo et al. (2013) |
|
Ehd4 |
|
SD/LD promotion |
3 |
Os03g0112700 |
LOC_Os03g02160 |
Zinc finger CCCH domain-containing protein |
Gao et al. (2013) |
|
Hd18 |
|
SD/LD promotion |
8 |
Os08g0143400 |
LOC_Os08g04780 |
Amine oxidase domain-containing protein |
Shibaya et al. (2016) |
1. SD: Short days; LD: Long days.
2. Locus ID of the Rice Annotation Project, National Agriculture and Food Research Organization.
3. Locus ID of the Rice Genome Annotation Project, Michigan State University.
4. Short references listed here are detailed with their full mention in Hori, Matsubara and Yano (2016).
Source: Adapted from Hori, K., K. Matsubara and M. Yano (2016), “Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics”, https://doi.org/10.1007/s00122-016-2773-5.
It has been found that the Hd1 protein can bind to the Ghd7 protein both in rice protoplasts and in cells from rice plants (Nemoto et al., 2016). This Hd1-Ghd7 complex may play an important role in repressing the Ehd1 (Early heading date 1) gene, a flowering promoter in rice, under long-day conditions. Conversely, the activation of Ehd1 under short-day conditions may not require Ghd7 function. Although most key functional natural variations identified in QTL genes have resulted in defective alleles, a specific mutation resulting in amino acid changes in the Hd17 (Heading date 17) gene was found to be beneficial for rice breeding as it improved Hd17 activity, reducing the amount of Ghd7 repressor mRNA (Matsubara et al., 2012). This selection occurred as a rare case during modern crossbreeding in rice.
Throughout the history of rice breeding, there are a few cases of cultivars that have progressed southward. A rare example of this is the major Chinese Taipei cultivar, Taichung 65, which has defective alleles in two heading date genes. One is a defective allele of the Hd1 gene, while the other is the Ehd1 gene; both of these defective alleles were introgressed from local landraces in Chinese Taipei into Japanese cultivar backgrounds to develop Taichung 65 (Doi et al., 2004; Wei et al., 2016). Here, both defective alleles of Hd1 and Ehd1 caused a late-flowering phenotype under short-day conditions. Thus, Taichung 65 possesses a long vegetative growth phase in subtropical areas of Chinese Taipei.
Itoh et al. (2018) evaluated the genetic contributions to the heading date of genome fragments from 10 distinct cultivars grown in various cultivation areas using 10 sets of chromosomal segment substitution lines (in total 429 lines). This work suggests that natural variations affecting heading date in various rice cultivars may be positioned at around 10-20 loci, although the same loci may have distinct heading date effects due to several distinct functional polymorphisms in a gene. In addition, many natural variations lead to neutral amino acid changes in genes or behave as silent mutations. Thus, phylogenetic trees tell the history (genetic distances) of genes and genomes but do not represent breeding selection due to functional changes of the target agricultural traits. Furthermore, some genomic regions contain clear signs of their past introgression events including selection for these heading date genes. These indicate complex genetic events for the heading date genes have been involved to establish each sub-species in rice.
Genetic factor affecting male sterility
Cytoplasmic male sterility (CMS) is a maternally inherited trait in which plants fail to produce functional pollen or anthers and is caused by interactions between the nuclei and mitochondria. A product of a CMS-causing gene encoded from the mitochondrial genome regulates nuclear genes via retrograde signalling, resulting in male sterility (reviewed in Fujii and Toriyama, 2008). However, a fertility restorer gene (Rf gene) in the nucleus genome suppresses the expression of the CMS-causing gene and recovers male fertility. A CMS line, a maintainer line and a fertility restorer line are thus often used for hybrid rice breeding and are known as a three-line system.
CMS plants are often obtained by successive backcrossing between distantly related species or sub‑species yielding cytoplasmic substitutions, although they are sometimes found in wild rice populations. Pollen abortion was observed in different developmental stages depending on the origins of the cytoplasm. For example, microspores abort just after meiosis in wild-abortive (WA)-type CMS, which is derived from wild rice in Hainan Island, whereas pollen aborts at a tricellular pollen stage in Boro (BT)-type CMS, which is derived from an indica rice variety Chinsurah Boro II (Table 4.5). In another case, exemplified by Chinese wild rice (CW)-type CMS, pollen looks morphologically normal but lacks the ability to germinate. WA‑type CMS is most widely used for female parents in hybrid rice breeding (reviewed in Huang et al., 2014). Other CMS types used for hybrid rice breeding include BT-type and Honglian (HL)-type CMS (reviewed in Huang et al., 2014).
Known CMS-causing genes from the mitochondrial genome are WA352 for WA-type CMS (Bentolila and Stefanov, 2012; Luo et al., 2013; Tang et al., 2017) and orf79 for BT-type CMS (Iwabuchi, Kyozuka and Shimamoto, 1993; Akagi et al., 1994; Kazama et al., 2016) (Table 4.5; reviewed in Huang et al., 2014; Kim and Zhang, 2018). WA352/orf352 and their sequence variants are reported in other CMS types such as D, DA, GA, ID, K (Luo et al., 2013), and RT102 (Okazaki et al., 2013). Orf79 and its sequence variants are reported in HL-type (Yi et al., 2002) and Lead rice (LD)-type CMS (Itabashi, Kazama and Toriyama, 2009; Table 4.5). WA352/orf352 is composed of parts from three genes of unknown function in the Nipponbare mitochondrial genome, namely orf284, orf224, and orf288, and a sequence of unknown origin (Luo et al., 2013; Okazaki et al., 2013). It is co-transcribed with rpl5, encoding ribosomal protein large subunit 5. The WA352 protein is reported to interact with a subunit of a respiration complex IV, resulting in reactive oxygen species (ROS) production and programmed cell death (PCD). Orf79 consists of a part of a coxI encoding cytochrome oxidase subunit I and has a sequence of unknown origin. It is co‑transcribed with atp6 encoding ATP synthase subunit 6 (Iwabuchi, Kyozuka and Shimamoto, 1993; Akagi et al., 1994; Kazama et al., 2016). ORFH79 of HL-CMS, which is encoded by a sequence variant of orf79, was reported to interact with a subunit of respiration complex III, resulting in ROS production and PCD leading to male sterility (Wang et al., 2013a).
Table 4.5. Type and characters of cytoplasmic male sterility (CMS)
|
CMS type |
Cytoplasm source |
Morphology of pollen1 |
Abortive stage |
CMS-associated gene |
Fertility restorer genes2 |
|---|---|---|---|---|---|
|
WA |
Wild rice with abortive pollen |
Unstained; irregular withered |
Early uninucleate microspore |
WA352 |
Rf3, Rf4 (=PPR782a) |
|
HL |
Wild rice (Hong Lian) |
Unstained; spherical |
Bicellular pollen |
orfH79 |
Rf5(='Rf1a),' Rf6 (PPR 894) |
|
BT |
Chinsurah Boro II (indica) |
Lightly stained; spherical |
Tricellular pollen |
orf79 |
Rf1a(='PPR791),' Rf1b (=PPR506) |
|
LD |
Lead rice (indica) |
Lightly stained; spherical |
Tricellular pollen |
L-orf79 |
Rf2 (glycine-rich protein) |
|
CW |
Wild rice (W1) |
Stained; round but no germination |
Germination |
orf307 |
Rf17(='retrograde-regulated' male sterility) |
1. pollen stainability with I2-KI.
2. the names of the PPR genes are based on the number of encoded amino acids.
Sources: Li, S., D. Yang and Y. Zhu (2007), “Characterization and use of male sterility in hybrid rice breeding”, https://doi.org/10.1111/j.1744-7909.2007.00513.x; Huang, J.Z. et al. (2014), “Workable male sterility systems for hybrid rice: Genetics, biochemistry, molecular biology, and utilization”, https://doi.org/10.1186/s12284-014-0013-6; Kim, Y.-J. and D. Zhang (2018), “Molecular control of male fertility for crop hybrid breeding”, https://doi.org/10.1016/j.tplants.2017.10.001.
Fertility Rf genes are present in the nuclear genome. Rf1 for BT-type CMS is present in chromosome 10 and acts gametophytically for fertility restoration. Rf3 and Rf4 are in chromosomes 1 and 10 respectively, and sporophytically restore fertility. Rf2 for LD-CMS has a weak restoration ability for BT-type CMS. There are some other Rf genes known to be responsible for weak fertility restoration (reviewed in Huang et al., 2014).
Molecular cloning has been performed for the following Rf genes: Rf1a and Rf1b for BT-type CMS (Kazama and Toriyama 2003; Komori et al., 2004; Akagi et al., 2004; Wang et al., 2006); Rf4 for WA-type CMS (Kazama and Toriyama 2014; Tang et al., 2014), and Rf5 (=Rf1) and Rf6 for HL-type CMS (Huang et al., 2015) (Table 4.5; reviewed in Huang et al., 2014; Kim and Zhang, 2018). These genes all encode pentatricopeptide repeat (PPR) proteins, which are known to be sequence-specific RNA-binding proteins (Table 4.5). These PPR proteins are targeted into the mitochondria and bind to orf79 or WA352-containing RNA, and promote RNA processing, such as RNA cleavage and degradation, resulting in the suppressed accumulation of products from CMS-causing genes. Rf2 encodes a glycine-rich protein, although its restoration mechanisms are unknown (Itabashi et al., 2011).
Thermo-sensitive genic male sterility (TGMS) and photoperiod-sensitive genic male sterility (PGMS) have also been used for hybrid rice breeding (reviewed in Huang et al., 2014). They are also referred to as environment-sensitive genic male sterility (EGMS). In these cases, a maintainer line is no longer necessary because male-sterile lines can be propagated through self-pollination under designated conditions. Hybrid seeds are produced by crossing between these male-sterile lines and any pollen parents. Thus, this method is called the two-line method. An example of this is the super hybrid rice “Liangyoupei9 (LYP9)” that was obtained using a P/TGMS line, Peiai64S (PA64S) and pollen parent 93-11. The TGMS and PGMS lines are sterile in high-temperature (typically >25°C) and long-day conditions (typically >14 h) but fertile in lower temperature and short-day conditions.
Although most genic male sterility is caused by loss-of-function alleles of genes that are essential for anther and pollen development (reviewed in Wang et al., 2013b), dominant genic male-sterile mutants have also been reported in rice and are expected to be useful for recurrent selection breeding to facilitate population improvements. The Pingxiang dominant male-sterile gene was designated Ms-p and mapped to chromosome 10 (Huang et al., 2007). The gene for the Sanming dominant male sterility was named SMS and mapped to chromosome 8 (Pang et al., 2017). The SMS dominant male-sterile line has been effectively used for recurrent selection breeding to obtain multiple abiotic stress-tolerant rice cultivars (Pang et al., 2017).
Genetic factors affecting sterility and weakness in hybridisation between cultivated species
Fitness reductions, such as lethality, weakness and sterility, are observed both in intraspecific and interspecific rice hybrids. This phenomenon is referred to as hybrid incompatibility. This subsection describes the hybrid incompatibility found in intraspecific hybrids of the Asian cultivated species O. sativa and in the interspecific hybrids between O. sativa and closely related species.
Hybrid compatibilities of the Oryza species with AA genomes (sativa complex) are governed by nuclear gene interactions and cytoplasm-nucleus gene interactions have also been detected. Details of the cytoplasmic male sterility genes have been described in the preceding section (“Genetic factor affecting male sterility”). Regarding nuclear genes involved in hybrid sterility, no locus common to natural mutations and induced mutations have been detected so far.
Hybrid sterility refers to the sterility of male gametes, female gametes or both gametes of F1 hybrids or hybrid progeny. Sterility can be sporophytic or gametophytic. Genetic studies of hybrid sterility have revealed two genetic models: i) allelic interactions at a single genetic locus (including tightly linked multiple genes) on heterozygotes; and ii) interactions at two independent genetic loci. In the allelic interaction model, selective abortion occurs depending on the genotype of the gametophyte but no sterility or other abnormal phenotype can be seen in either homozygote. In the intraspecific crosses of O. sativa, many genes corresponding to the allelic interaction types are reported, such as Sa (Zhuang et al., 1999), Sc (Zhuang et al., 2002), S24 (Kubo et al., 2008), S25 (Win et al., 2009), S35 (Kubo et al., 2008) as male sterility genes and S5 (Ikehashi and Araki, 1986) and S7 (Yanagihara, Kato and Ikehashi, 1992) as female sterility genes. The cross combination of O. rufipogon and O. sativa, S36 (Win et al., 2009) and ESA1 (Hou et al., 2019) was found to cause hybrid sterility.
Some cases of hybrid sterility are governed by intergenic interactions at two or more loci. In intraspecific hybrids of the cultivated species, DPL1/DPL2 genes for gametophytic pollen sterility (Mizuta, Harushima and Kurata, 2010) and HSA1, HSA2, and HSA3 (Kubo and Yoshimura, 2005) for sporophytic embryo sac sterility have been reported. DGS1/DGS2 is known for interspecific hybridisation between O. sativa and O. nivara (Nguyen et al., 2017).
More than 40 causal loci/QTL for hybrid sterility have been reported. These reported genes mainly consist of allelic interaction type genes. Incompatible genotypes of the sporophyte or gametophyte determine sterility. Out of these reported gene loci, 11 genes have been isolated and characterised (Table 4.6). Cloning studies have revealed that the allelic interaction type loci are composed of two or more genes encoding different protein families or a tandem duplication of gene copies. The causal genes of hybrid sterility do not likely function in a single or specific physiological pathway essential for gamete development. However, genes encoding proteinases or peptidases have often been found to be the causal molecules.
Table 4.6. Cloned genes affecting hybrid sterility in intraspecific crosses of O. sativa L.
|
Mendelian locus |
Chr. |
Affected gametophyte |
Gene |
Gene function |
Reference |
|---|---|---|---|---|---|
|
S5 |
6 |
Female |
ORF3 |
Heat shock protein Hsp70 |
Yang et al. (2010) |
|
ORF4 |
Unknown protein with transmembrane region |
Yang et al. (2010) |
|||
|
ORF5 |
Eukaryotic aspartic proteases |
Chen et al. (2008) |
|||
|
S7 |
7 |
Female |
ORF3 |
Tetratricopeptide repeat domain-containing protein |
Yu et al. (2016) |
|
S-a |
1 |
Male |
SaF |
F-Box Protein |
Long et al. (2008) |
|
SaM |
SUMO E3 Ligase-like Protein |
Long et al. (2008) |
|||
|
S-c |
3 |
Male |
S-c |
DUF1618 domain-containing protein |
Shen et al. (2017) |
|
DPL1 |
1 |
Male |
DPL1 |
Unknown protein |
Mizuta, Harushima and Kurata (2010) |
|
DPL2 |
6 |
Male |
DPL2 |
Unknown protein |
Mizuta, Harushima and Kurata (2010) |
|
hsa1 |
12 |
Female |
HSA1a |
DUF1618 domain-containing protein |
Kubo et al. (2016) |
|
HSA1b |
Uncharacterised protein |
Kubo et al. (2016) |
Sources: Chen, X.-P, et al. (2008), “Ammonia-oxidizing archaea: Important players in paddy rhizosphere soil?”, https://doi.org/10.1111/j.1462-2920.2008.01613.x; Kubo, T. et al. (2016), “Two tightly linked genes at the hsa1 locus cause both F1 and F2 hybrid sterility in rice”, https://doi.org/10.1016/j.molp.2015.09.014; Long, Y. et al. (2008), “Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes”, https://doi.org/10.1073/pnas.0810108105; Mizuta, Y., Y. Harushima and N. Kurata (2010), “Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes”, https://doi.org/10.1073/pnas.1003124107; Shen, R. et al. (2017), “Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice”, https://doi.org/10.1038/s41467-017-01400-y; Yang, J. et al. (2010), “A killer-protector system regulates both hybrid sterility and segregation distortion in rice”, Science, Vol. 337, pp. 1336-1340; Yu, Y. et al. (2016), “Hybrid sterility in rice (Oryza sativa L.) involves the tetratricopeptide repeat domain containing protein”, https://doi.org/10.1534/genetics.115.183848.
The gametophytic sterility genes cause skewed segregation in the progeny of the heterozygous hybrid due to their allelic interactions. This phenomenon is also called transmission ratio distortion (TRD). Both homozygotes in the progeny of the heterozygous plant do not cause remarkable phenotypes, including sterility. The positively selected alleles are expected to penetrate the population at a faster rate than normal Mendelian factors in the heterozygous population.
Some local varieties or wild species harbouring neutral alleles have been found to be compatible with any allelic type (Chen et al., 2008). Neutral alleles are in widespread use in crossbreeding and breeding programmes for the F1 hybrids in some Asian countries (Chen et al., 2008). New Rice for Africa (NERICA), which is a hybrid cultivar derived from an interspecific hybrid between O. sativa and African cultivated species O. glaberrima, has been widely grown in Africa. The potential opportunities of hybridisation with O. sativa cultivars are increasing. Generally, the hybrid between O. sativa and O. glaberrima does not produce self-pollinated seeds due to complete pollen sterility. Many genes for hybrid sterility are reported in the O. sativa/O. glaberrima cross: S1 (Sano, 1990), S18 (Doi, Taguchi and Yoshimura, 1998), S19 (Taguchi, Doi and Yoshimura, 1999; Zhang et al., 2011), S20, S21 (Doi, Taguchi and Yoshimura, 1999), S29 (Hu et al., 2006), S33 (Ren et al., 2005), S34 (t) (Zhang et al., 2002), S36 (Li et al., 2011) and S37, S38, S39 (Xu et al., 2014). S1 causes both pollen and seed sterility but the other genes cause only pollen sterility.
The hybrid weakness among the O. sativa/O. rufipogon gene pool is genetically divided into two classes, hybrid weakness or lethality found in the F1 generation (F1 hybrid weakness/lethality) and those found in the F2 and subsequent generations (F2 hybrid weakness/lethality or hybrid breakdown). Hwa, Hwc and Hwi1 are reported as F1 hybrid weakness genes (Kuboyama et al., 2009; Ichitani et al., 2011; Chen et al., 2014). Duplicate recessive genes such as hwb1/hwb2 (Oka, 1957), hwd1/hwd2 (Fukuoka, Namai and Okuno, 1998) and hwe1/hwe2 (Kubo and Yoshimura, 2002) are known as causal genes for F2 hybrid weakness. Further analyses of Hbd2/Hbd3 (Yamamoto et al., 2007; Yamamoto, 2010), Hwi1/Hwi2 (Chen et al., 2014) and Hwc3 (Nadir et al., 2019) revealed that deleterious interactions between these genes cause an autoimmune response.
Breeding approaches
Rice breeding has been supported by a variety of breeding techniques based on accumulated research and traditional experience over many years. Traditional breeding methods include the collection and evaluation of genetic resources, induction of artificial mutations and the selection of individuals and lines. It is described in detail in the Kaneda publication (1993).
To achieve stable rice production, high yields, lodging resistance, resistance to pests and disease, tolerance to abiotic stress such as high and low temperature, drought, salinity as well as good eating quality and health functionality are the main targets for rice breeding programmes. With temperature increases due to global warming, responses to high-temperature damage such as reductions in yield, grain quality and sterility need to be improved. In Japan and Korea, rice has been used as a feed crop, as whole crop silage or grain. High biomass and digestibility by animals are target traits in breeding programmes.
The lodging resistant variety IR8 was the first variety developed with a semi-dwarfing gene, sd1, and contributed greatly to the Green Revolution in the 1960s (Khush, Coffman and Beachell, 2001). Since then, the development of semi-dwarfing varieties has been the main goal in most rice-producing countries. It was revealed that the Sd1 gene encodes GA20-oxidase (Os20ox2) and that the short stature phenotype was caused by a loss-of-function sd1 (Sasaki et al., 2002). Interestingly, different types of the Sd1 alleles, which showed weak function, have independently been artificially induced in Japan and the United States. These several types of sd1 alleles have been used to develop new varieties (Sasaki et al., 2002). Major targets of disease resistance in rice breeding are rice blast, bacterial leaf blight, brown spot, sheath blight, rice stripe, rice dwarf and yellow dwarf, and, for pest resistance, they are brown planthopper, green leafhopper, rice stem borer and pecky rice bug (Annex 4.A and Annex 4.B).
Crossbreeding and selection are standard methods in rice breeding programmes. In the 1960s, since the cytoplasmic male sterility and its restorer genes became available, the development of F1 hybrid varieties began and their commercial production increased (Cheng et al., 2007; FAORAP and APSA, 2014; Xie and Zhang, 2018).
Initially, a three-line system (cytoplasmic male-sterile [CMS], maintainer and restorer lines) was employed to develop F1 hybrid cultivars. However, later strategies involved male parents for two-line hybrids based on thermo- or photo-sensitive male sterile lines to enhance the effective F1 seed production (FAORAP and APSA, 2014; Cheng et al., 2007). In Asia, following the success of producing F1 rice hybrids China, several other countries such as Bangladesh, India, Indonesia, Myanmar, the Philippines and Viet Nam introduced the development and production of F1 hybrids. The level of heterosis has been clear in indica and japonica crosses but a relatively small level of heterosis was observed between japonica crosses. This poor heterosis resulted in limitations for the F1 hybrid cultivars in Japan and Korea.
To improve a particular trait of interest, induced mutations and marker-assisted selection (MAS) have been utilised. So far, lodging resistance, disease resistance and changes to chemical components in the endosperm have been achieved through the selection of mutants induced by gamma-ray and chemical mutagens (Rutger, 1992; Nakagawa and Kato, 2017).
Due to the progress in genome sequencing and methods for genetic analysis, QTL identification and cloning have been routinely performed (Yano, 2001; Yamamoto, Yonemaru and Yano, 2009). In association with the dramatic progress in the detection of sequence variations, MAS has already been an effective method for rice breeding programmes (Jena and Mackill, 2008; Cobb, Biswas and Platten, 2019).
The utilisation of biotechnology in rice breeding started with transformation technologies in the late 1980s. The use of developed genome editing technologies has been promoted since 2010 (Christian et al., 2010). Since the CRISPR/Cas9 system was published in 2012 (Gasiunas et al., 2012; Jinek et al., 2012), however, genome editing technologies have rapidly spread, and genome editing for rice was developed in 2013 utilising the CRISPR/Cas9 system (Annex 4.D).
Hybridisation and introgression
Outcrossing and gene flow in rice
Cultivated rice is a strictly self-pollinated species. However, cross-pollination and gene flow can occur if rice is growing in the vicinity of weedy rice or other AA genome wild species, which show some degree of sterility and whose flowers remain open at the time of pollination. Oka (1988a) reported that the natural crossing frequency of japonica ranges from 0.6% to 3.9% and that of indica ranges from 0.0% to 6.8% (Table 4.7). The natural crossing rate of wild rice is greater than that of cultivated rice. There are some wild rice varieties with crossing rates greater than 50% (Table 4.7). This variation could be due to different growing conditions, for example the distance between rice and weedy rice/wild species, wind speed, opening of flower, stigma protrusion or the degree of sterility of the weedy rice/wild species, thus allowing for open pollination.
Table 4.7. Outcrossing rates estimated in wild and cultivated rice species by different methods
|
Taxa/type |
Origin |
Method |
No. of populations |
Outcrossing (%) |
Reference1 |
|---|---|---|---|---|---|
|
Asian O. perennis Perennial |
Chinese Taipei |
Marker gene |
1 |
30.7 |
Oka (1956c) |
|
Thailand |
Marker gene |
1 |
44 |
Oka and Chang (1961) |
|
|
Thailand |
Isozyme markers |
1 (NE88) |
50.6 |
Barbier (1987) |
|
|
Intermediate |
Thailand |
Isozyme markers |
1 (CP20) |
55.9 |
Barbier (1987) |
|
Perennial |
India |
Variance ratio |
1 |
37.4 |
Oka and Chang (1959) |
|
Sri Lanka |
Variance ratio |
2 |
22.4-26.5 |
Sakai and Narise (1959) |
|
|
Annual |
India |
Variance ratio |
1 |
21.7 |
Oka and Chang (1959) |
|
India |
Variance ratio |
3 |
16.6-33.9 |
Sakai and Narise (1960) |
|
|
India |
Marker gene |
1 |
7.9 |
Roy (1921) |
|
|
Thailand |
Isozyme markers |
1 (NE4) |
7.2 |
Barbier (1987) |
|
|
Weedy |
India |
Variance ratio |
2 |
17.3-20.6 |
Oka and Chang (1959) |
|
breviligulata |
Africa |
Variance ratio |
2 |
3.2-19.7 |
Morishima et al. (1963) |
|
sativa |
India |
Marker gene |
34 |
0-6.8 |
Butany (1957) |
|
indica |
Africa |
Marker gene |
2 |
0-1.1 |
Roberts et al. (1961) |
|
indica |
Chinese Taipei |
Marker gene |
4 |
0.1-0.3 |
Oka (unpublished) |
|
japonica |
Chinese Taipei |
Marker gene |
5 |
0.6-3.9 |
Oka (unpublished) |
|
indica |
Sri Lanka |
Variance ratio |
1 |
3.6 |
Sakai and Narise (1960) |
1. Short references listed here are detailed with their full mention in Oka (1988a).
Source: Oka, H.-I. (1988a), “Ancestors of cultivated rice”, in Origin of Cultivated Rice, Japan SSP, Tokyo/Elsevier, Amsterdam, pp. 18-22.
Wild relatives show outcrossing to a varying degree. In several wild species or weedy rice species, the anthers are long with extruded stigma, favouring outcrossing. The Asian forms of O. perennis complex showed outcrossing ranging from 7.0% to 55.9%, which was higher in perennial than annual types. Outcrossing is dependent on flower morphology, stigma exertion, male sterility, the duration of flower opening and other environmental factors (Endo et al., 2009).
Outcrossing is also affected by the capacity of the stigma to receive alien pollen before self-pollination and the capacity of anthers to emit pollen to pollinate other plants in their proximity. Intervals from flowering to pollen emission, stigma size and extrusion of the stigmas from the flower are the other factors affecting outcrossing.
Lu, Yang and Ellstrand (2016) summarised the results of different studies conducted in China, Costa Rica, Korea, Spain and the US on pollen-mediated gene flow from transgenic to non-transgenic rice. Outcrossing was determined using molecular marker analysis. The gene flow frequency ranged from 0.0% to 0.47% except in one study where it ranged from 1.0% to 2.3% (Table 4.8).
Several studies have shown that the strictly self-fertilising nature and short life of the pollen grains of cultivated rice plants account for the extremely low gene flow from transgenic rice to other non-rice cultivars. However, through pollen-mediated gene flow, transgenes can move from cultivated rice to nearby weedy rice (O. sativa f. spontanea) or any of the six wild species (O. rufipogon, O. nivara, O. breviligulata (O. barthii), O. longistaminata, O. meridionalis, O. glumaepatula) belonging to sativa complex growing sympatrically or as intermixed populations.
Several studies have shown that the outcrossing of rice with weedy rice and AA genome wild species of rice, occurs in field conditions in natural habitats. However, it is not known precisely how fitness-enhancing transgenes will accumulate in these populations and how far these will have unwanted environmental consequences. The risks could be assessed by: i) estimating transgene frequencies; ii) assessing the expression levels of transgenes in wild populations; and iii) measuring the fitness change.
Rong et al. (2012) grew 3 genetically engineered (GE) insect-resistant lines with non-transgenic lines at four scales ranging from 9 m² to 576 m2 (8 GE: 1 non-GE). Out of 1.3 million seeds examined from non‑GE rice plots, very low frequencies of the transgene were detected (<0.1%). Chen et al. (2004) estimated outcrossing rate from cultivated to weedy rice (0.011-0.046%) and from cultivated to wild rice (1.21-2.19%). Thus, transgenes can be expressed in weedy rice and wild species and potentially alter the fitness of the wild/weedy plants and the dynamics of the wild population.
Table 4.8. Field experiments to detect the frequency of pollen-mediated (trans)gene flow from cultivated rice to weedy rice
|
Crop |
(Trans)gene |
Location |
Marker used to detect gene flow |
Observed gene flow frequency (%) |
References1 |
|---|---|---|---|---|---|
|
Glufosinate-resistant rice |
United States |
Glufosinate-resistant marker |
0 |
Sanders et al. (1998) |
|
|
Imidazolinone-resistant rice |
United States |
Imidazolinone-resistant marker |
0.00 |
Sanders et al. (2000) |
|
|
Imidazolinone-resistant rice line ‘CL 2551’ |
United States |
Imidazolinone-resistant marker and SSR molecular fingerprinting |
0.0-0.05 |
Estorninos et al. (2002) |
|
|
GE rice |
gusA and bar gene |
Spain |
Glucuronidase marker |
0.036±0.006 |
Messeguer et al. (2004) |
|
GE rice (Nam29/TR18) |
bar gene |
South Korea |
Basta-resistance marker |
0.011-0.046 |
Chen et al. (2004) |
|
Imidazolinone-resistant Clearfield® rice |
United States |
Imidazolinone-resistant marker and SSR molecular fingerprinting |
0.003-0.008 |
Shivrain et al. (2007) |
|
|
GE rice |
PPT-R |
Costa Rica |
Glufosinate-resistant marker |
1.0-2.3 |
Olguin et al. (2009) |
|
GE rice |
Protox (protoporphyrino-gen oxidase) gene |
South Korea |
PPO-resistance marker |
0.04 |
Chun et al. (2011) |
|
GE rice |
bar gene |
China |
Basta-resistance marker |
0.002-0.342 and 0.090 |
Jia et al. (2014) |
|
indica and tropical japonica rice cultivars |
United States |
SSR molecular fingerprinting |
0 |
Gealy et al. (2015) |
|
|
GE rice (Xiang 125S/Bar68-1) |
bar gene |
China |
Glufosinate-resistant marker |
0.395-0.470 and 0‑0.187 |
Sun et al. (2015) |
1. Short references listed here are detailed with full mention in Lu, Yang and Ellstrand (2016).
2. Non-transgenic variety
Source: Lu, B.R., X. Yang and N.C. Ellstrand (2016), “Fitness correlates of crop transgene flow into weedy populations: a case study of weedy rice in China and other examples”, https://doi.org/10.1111/eva.12377.
Experimental production of interspecific hybrids
A number of studies have been conducted over the years on interspecific crosses for cytogenetic research involving genome analysis, chromosome pairing analysis in F1 hybrids and more recently on the introgression of useful genes from wild species into cultivated rice for tolerance to biotic and abiotic stresses, diversification of cytoplasmic male sterility sources and to introgress QTLs or yield-enhancing loci “wild species alleles” (Brar and Singh, 2011; Brar and Khush, 2018). Hybrids have been successfully produced through crosses made between rice and all of the 22 wild species of Oryza except O. schlechteri. Several crossability barriers limit the transfer of genes from wild species into rice (Sitch, 1990; Khush and Brar, 1992). Nezu, Katayama and Kihara (1960) studied crossability and chromosome affinity among 17 species of Oryza and found that crossability differs in different cross-combinations.
Crossability amongst AA genome species is relatively high and crosses of rice with the six diploid wild species of the sativa complex (2n = 24, AA) can be made easily. These hybrids have been produced through direct crosses (without embryo rescue) of rice with all of the species of the sativa complex. Plant breeders make these crosses routinely by crossing elite breeding lines of rice (O. sativa) with the wild species accessions possessing the genes for the target agronomic traits. F1 offspring are partially fertile and these are either selfed or backcrossed with the recurrent rice parent to develop elite breeding lines for the introgression of useful genes from wild species. Several institutes have produced a series of interspecific hybrids (rice × wild species) and introgression lines for cytogenetic and breeding research.
In natural conditions, where rice and diploid wild species of the sativa complex grow sympatrically, cross-hybridisation occurs frequently, resulting in the production of interspecific hybrids, intermediate progenies or hybrid swarms. Such types of cross-hybridisation in natural habitats are common among rice, weedy rice and AA genome wild species.
Unlike the AA genome wild species, hybrids cannot be produced through direct crosses between rice and wild species belonging to the officinalis complex (BB, CC, BBCC, CCDD, EE genomes) without embryo rescue in the F1. No report is available on the natural crossing and production of hybrids between rice and species of this complex. Hybrids have been produced between rice and wild species of the officinalis complex through embryo rescue of developing F1 seeds (Jena and Khush, 1990; Brar, Elloran and Khush, 1991). In one experiment, a total of 26 034 spikelets of three lines of O. sativa were pollinated with the CC genome wild species (O. officinalis) and the seed set ranged from 8.82% to 17.30% (Jena and Khush, 1990). From these F1 seeds, embryos were rescued after 14 days of pollination and cultured on MS medium. While the germination ratios for the embryos were high (from 56.8% to 70.0%), the rate of plant survival after culture was lower. As a result, crossability (number of hybrid plants obtained/total number of spikelets pollinated) ranged from 1.0% to 2.3%.
Crosses of O. sativa with the CCDD genome species (O. latifolia) were made by Multani et al. (2003). The seed set was 19.8%, germination of the hybrid embryos was 85.5%, with crossability being 7.6%. In the BC1, 10 144 spikelets of F1, were pollinated with rice pollen and crossability was 0.11%, similarly in the BC2 and BC3 crossability was 0.21% and 0.62% respectively.
Multani et al. (1994) made crosses among rice and the EE genome wild species (O. australiensis). Seed set ranged from 2.3% to 2.9%. Although embryos germinated well in the culture medium (50.4‑62.9%), however, crossability was extremely low (0.25-0.90%). Data on such a low crossability in controlled crosses of rice and distantly related wild species support the lack of any report of hybrids under natural field conditions. Low crossability and other barriers may be the reason why no natural hybrids exist between rice and wild species, except for the AA genome species.
A number of genes for several agronomic traits, brown planthopper (BPH) resistance, bacterial blight (BB) resistance and blast resistance, have been introgressed from wild species of the sativa complex into cultivated rice and improved varieties have been released for commercial cultivation (Brar and Khush, 1997, 2018; Table 4.9). Among the classical examples are the introgression of a gene for grassy stunt virus resistance from O. nivara to cultivated rice varieties (Khush, 1977) and the transfer of a cytoplasmic male-sterile (CMS) source from wild rice, O. sativa f spontanea (Lin and Yuan, 1980).
Other useful genes from wild species such as Xa21, Xa23 and Xa38 for BB resistance have been introgressed into rice. Xa21 has a broad spectrum of resistance and has been pyramided along with other genes for BB resistance (Singh et al., 2001). Many varieties have been released through marker-assisted selection (MAS) using Xa21 and other stacked genes. Genes for tolerance to tungro and tolerance to acid sulphate soil conditions have been transferred from O. rufipogon into the indica rice cultivar (Table 4.9). Recently, at Punjab Agricultural University, India, Xa38 and xa45(t) have been identified from O. nivara and O. glaberrima respectively for resistance to BB.
Advanced breeding lines carrying these genes have been developed and one of the lines (PR127) carrying xa45(t) has been released for commercial cultivation. Furthermore, many introgression lines harbouring variations for yield component traits from the five different wild species with AA genomes have been developed (Bhatia et al., 2017). The African rice (O. glaberrima) has been used extensively by the Africa Rice Center and a number of indica rice varieties NERICA have been released with introgressed genes for early heading, weed competitive ability and tolerance to biotic and abiotic stress.
Despite limited recombination between chromosomes of rice and wild species such as O. officinalis, O. minuta, O. latifolia, O. australiensis, and O. grandiglumis of the officinalis complex, some genes for resistance to BPH, BB, blast, and whitebacked planthopper (WBPH) have been successfully introgressed into rice (Table 4.9). Some varieties have also been released commercially.
Table 4.9. Introgression of genes from wild Oryza species into cultivated rice
|
Trait(s) transferred into O. sativa (AA) |
Wild Species (donor) |
Gene/QTLs |
Genome |
|---|---|---|---|
|
Grassy stunt resistance |
O. nivara |
Gs |
AA |
|
Bacterial blight resistance |
O. longistaminata |
Xa21 |
AA |
|
O. rufipogon |
Xa23 |
AA |
|
|
O. nivara |
Xa38 |
AA |
|
|
O. officinalis |
Xa 29(t) |
CC |
|
|
O. minuta |
Xa27 |
BBCC |
|
|
O. latifolia |
Unknown |
CCDD |
|
|
O. australiensis |
Unknown |
EE |
|
|
O. brachyantha |
Unknown |
FF |
|
|
O. glaberrima |
xa45(t) |
AA |
|
|
Blast resistance |
O. glaberrima |
Unknown |
AA |
|
O. rufipogon |
Unknown |
AA |
|
|
O. nivara |
Unknown |
AA |
|
|
O. glumaepatula |
Unknown |
AA |
|
|
O. barthii |
Unknown |
AA |
|
|
O. minuta |
Pi9 |
BBCC |
|
|
O. australiensis |
Pi40 |
EE |
|
|
Brown planthopper (BPH) resistance |
O. rufipogon |
Bph35 |
AA |
|
O. nivara |
Bph34 |
AA |
|
|
O. officinalis |
Bph11, Bph12, Bph14, Bph15 |
CC |
|
|
O. eichingeri |
Bph13 |
CC |
|
|
O. minuta |
Bph20, Bph21 |
BBCC |
|
|
O. latifolia |
Unknown |
CCDD |
|
|
O. australiensis |
Bph10, Bph18 |
EE |
|
|
Whitebacked planthopper (WBPH) resistance |
O. officinalis |
Wbph7(t), Wbph8(t) |
CC |
|
O. latifolia |
Unknown |
CCDD |
|
|
Cytoplasmic male sterility (CMS) |
O. sativa f spontanea |
Wild abortive (WA) |
AA |
|
O. perennis |
Unknown |
AA |
|
|
O. glumaepatula |
Unknown |
AA |
|
|
O. rufipogon |
Unknown |
AA |
|
|
Tungro tolerance |
O. rufipogon |
Unknown |
AA |
|
Tolerance to iron toxicity |
O. rufipogon |
Unknown |
AA |
|
O. glaberrima |
Unknown |
AA |
|
|
Drought-related traits |
O. glaberrima |
QTL |
AA |
|
Tolerance to aluminium toxicity |
O. rufipogon |
QTL |
AA |
|
Tolerance to acidic conditions |
O. glaberrima |
Unknown |
AA |
|
O. rufipogon |
Unknown |
AA |
|
|
Tolerance to phosphorus deficiency |
O. glaberrima |
Unknown |
AA |
|
O. rufipogon |
Unknown |
AA |
|
|
Yield-enhancing loci (wild species alleles) |
O. rufipogon |
QTL |
AA |
|
O. nivara |
QTL |
AA |
|
|
O. grandiglumis |
QTL |
CCDD |
|
|
Earliness, stress tolerance, weed competitive ability |
O. glaberrima |
Unknown |
AA |
|
Increased elongation ability |
O. rufipogon |
Unknown |
AA |
Note: QTL – Quantitative trait locus.
Source: Khush, G.S. and D.S. Brar (2017), “Alien introgression in rice”, https://doi.org/10.1007/s13237-017-0222-7.
Introgression from the CC genome species: Several introgression lines have been produced from the crosses of O. sativa and O. officinalis (Jena and Khush, 1990). Genes for resistance to BPH, e.g. Bph10, bph11, bph12 and Bph18, and two QTL, qBph1, qBph2 and Xa29(t) for BB resistance have been introgressed into the progenies. Four breeding lines have been released as varieties (MTL95, MTL98, MTL103, and MTL110) for commercial cultivation in the Mekong Delta of Viet Nam.
Introgression from the BBCC genome species: Interspecific hybrids have been produced between O. sativa and the tetraploid wild species O. minuta (BBCC). Advanced introgression lines were produced using the embryo rescue of F1 hybrids followed by backcrossing with the O. sativa parent (Brar et al., 1996). Genes for resistance to BB and blast have been introgressed into rice. Blast resistance gene (Pi9) has a wide spectrum resistance and has been used in breeding programmes in India. Two genes (Bph20 and Bph21) for BPH resistance have been introgressed from O. minuta into rice.
Introgression from the CCDD genome species: Previous investigations have developed hybrids between rice and O. latifolia (CCDD) (Sitch, 1990; Brar, Elloran and Khush, 1991). Several introgression lines derived from this cross have been evaluated for the introgression of useful traits (Multani et al., 2003). Ten allozymes of O. latifolia, such as Est5, Amp1, Pgi1, Mdh3, Pgi2. Amp3, Pgd2, Est9, Amp2 and Sdh1, located on 8 of the 12 chromosomes were observed in the introgression lines. Alien introgression was also detected for morphological traits such as long awns, earliness, black hull, purple stigma and apiculus. Genes for resistance to BB, BPH and WBPH have been introgressed into elite breeding lines from O. latifolia. Yield-enhancing loci in the population derived from crosses of the japonica cultivar Hwaseongbyeo × O. grandiglumis (CCDD) have been identified. Of the 39 QTL, O. grandiglumis contributed desirable alleles in 18 QTL.
Introgression from EE genome species: Hybrids between cultivated rice and the EE genome species O. australiensis were produced (Multani et al., 1994). Of the 600 BC2F4 progenies, four were resistant to BPH. Introgression was observed for morphological traits such as long awns and earliness and Amp3 and Est2 allozymes. Resistance to BPH was found to be under monogenic recessive control in two progenies and a dominant gene conveyed resistance in the other two. The dominant gene in one of the progenies designated as Bph10 conferred resistance to three biotypes of BPH in the Philippines. Marker RG457 detected introgression from O. australiensis. Co‑segregation for the BPH reaction and molecular markers showed a gene for BPH resistance linked to RG457, with a distance of 3.68 centimorgan (cM) (Ishii et al., 1994). Introgression was detected for two other genes from O. australiensis: Bph18 for BPH resistance and a major gene Pi40 (t) for blast resistance.
Introgression from FF genome species: A hybrid between cultivated rice and the FF genome species O. brachyantha was produced and 149 backcross progenies were obtained. Introgression was obtained for resistance to Philippines bacterial blight races 1, 4, and 6 (Brar et al., 1996). Gene transfer in the introgression lines was not associated with any undesirable traits of O. brachyantha.
Introgression from KKLL genome species: To introduce salt tolerance into cultivated rice, a hybrid of cultivated rice and O. coarctata Roxb (KKLL genome) was produced by embryo rescue method (Jena, 1994). Although salt tolerance level has not been evaluated, viable hybrid plants showed triploid nature and possessed several phenotypic characteristics resembling O. coarctata.
Information and data on natural introgression
Gene flow from cultivated rice to wild rice under experimental field conditions
Under experimental field conditions, gene flow from cultivated rice (O. sativa) to wild rice (O. rufipogon) was confirmed using simple sequence repeat (SSR) markers specific to cultivated rice (Song et al., 2003). Of the 23 776 seedlings from O. rufipogon, 294 were identified to be interspecific hybrids between O. sativa and O. rufipogon. The frequency of the gene flow significantly decreased with distance from the pollen sources of the cultivated rice. The maximum observed distance of the gene flow was 43.2 m.
Introgression from cultivated rice to wild rice under natural field conditions
Under natural field conditions, gene flow also occurs from cultivated rice to wild rice populations. In Thailand, 7 out of 13 wild rice populations were found to have glutinous genes specific to cultivated rice (Oka and Chang, 1961). Most of them seemed to be caused by occasional gene flow from cultivated rice but one maintained high frequency (28.3%) of the glutinous gene in these populations. This population may have survived beyond the initial hybrid generation with a large amount of genetic variability through introgression. In southern China, genetic variation among six wild populations in Guangdong Province was surveyed using SSR markers (Zhu et al., 2017; Jin et al., 2017). Of these, one population spatially close to rice fields showed less genetic differentiation from the local cultivated rice groups, indicating that introgression from cultivated rice considerably altered the genetic structure of the wild population.
Introgression from cultivated rice to weedy rice
Weedy rice is a conspecific form of cultivated rice. Some weedy groups seem to have evolved from cultivated ancestors according to whole-genome sequence analyses (Li et al., 2017) and others have been generated by gene flow between cultivated and wild rice in tropical Asia (Pusadee et al., 2013, Song et al., 2014). Under experimental field conditions, gene flow from cultivated rice to weedy rice was estimated to be about 0.036% when there was a 25 cm distance between the plants (Messeguer et al., 2004).
In the United States, herbicide-resistant rice varieties that were a result of mutation breeding were first marketed in 2001 (Tan et al., 2005). Low levels of natural hybrids were initially reported between resistant varieties and weedy rice (Shivrain et al., 2007, 2008). However, weed control using the herbicide has forced strong selection on weedy rice populations. In 2010, resistant weedy rice plants were detected in all 26 fields with a history of herbicide-resistant varieties in Arkansas (Burgos et al., 2014). Although most weedy rice offspring (63%) were still sensitive to the herbicide, introgression of resistant alleles to the weedy rice population, by outcrossing between weedy rice and the herbicide-resistant varieties, is highly likely to be ongoing.
Various interactions with other organisms (ecology)
Interactions in natural ecosystems and agroecosystems
Interaction with pests
Interaction with vertebrate pests
Various birds, such as sparrows, crows, pigeons, parrots, weaverbirds and ducks feed on rice around the world. Damage by birds occurs during the sowing and harvest periods. In Japan, rice crops were damaged mainly by tree sparrows, jungle crows, carrion crows, Oriental turtle doves and spot-billed ducks (Lane, Azuma and Higuchi, 1998; Fujioka and Yoshida, 2001). However, the ears of the harvested rice and gleanings were fed on mostly by ducks, geese and cranes in the winter (Shimada 2002; Fujioka et al., 2010). The Japanese Red List 2020 made the following designations: greater white-fronted geese (near threatened, NT), bean geese (vulnerable species, VU), hooded cranes (VU) and white-naped cranes (VU). Cackling geese (critically endangered, CR), snow geese (CR) and lesser white-fronted geese (endangered species, EN) were included, although there have been few arrivals (Fujioka et al., 2010).
Interaction with invertebrate pests
Many insect pests have been reported in rice cultivation areas around the world. Grist and Lever (1969) lists more than 800 insect pests but only approximately 20 species are usually important in tropical Asia (Dale, 1994). In China, it has been reported that 347 species of insects infest rice plants, of which 74 species cause economic damage, 5 species cause serious damage and 31 species cause problems depending on the region and year (Zhang, 1992).
In Japan, 232 species of insects have been reported to infest rice plants (Japanese Society of Applied Entomology and Zoology, 2006) and 8 species and 1 group (rice bugs) have been designated as Specified Pests by the Ministry of Agriculture, Forestry and Fisheries, Japan. In West Africa, 330 species of insects have been collected from paddy fields but only about 10 species are of major importance (Heinrichs and Barrion, 2004). In India, 71 species of insects have been observed in paddy fields, including root feeders, stem borers, defoliators, grain suckers, leafhoppers and plant hoppers (Ane and Hussain, 2015).
The major pest species that damage rice not only vary from region to region but also vary from year to year in the same region and by rice growth stage. The characteristics of pests are described below in terms of their feeding habits, host ranges and migration. The detailed classification and ecology of insect pests of rice plants have been previously described (Grist and Lever, 1969, Ressing et al., 1986, Khan et al., 1990; Pathak and Khan, 1994; Heinrichs, 1994; Heinrichs and Barrion, 2004).
Feeding habit
Stem borers: Fifty species have been reported worldwide, most of them in the order Lepidoptera (family: Crambidae, Noctuidae and Pyralidae) (Khan et al., 1990). The larvae infest widely from seedling to maturing stage of rice plant. Larvae penetrate the leaf sheath and stem of the rice, causing leaf death (dead heart), and no filling of the spikelets (white head). The host range varies greatly from monophagy and oligophagy to polyphagy, depending on the species (Khan et al., 1990). Major species reported are Chilo suppressalis and Scirpophaga incertulas, Scirpophaga innotata, Sesamia inferens in Asia and Oceania, C. partellus, C. diffusilineus, Maliarpha separatella in Africa (Pathak and Khan, 1994). C. suppressalis was one of the most important paddy rice pests in Japan but it decreased its number and infestation area rapidly from the 1960s, and damage from this pest is now hardly reported. The decrease in C. suppressalis is largely attributable to changes in rice varieties, earlier transplanting and the introduction of harvesting machines (Kiritani, 2007).
Stalk-eyed flies (Diptera: Diopsidae) are reported as stem borers only in Africa. The larvae penetrate the stem and produce a dead heart. Khan et al. (1990) reported five species but Diopsis macrophthalma (= D. longicornis) and D. indica (= D. apicalis) are considered the main species.
Foliage feeders: Lepidopteran insect larvae, such as armyworm, cutworm, rice green semilooper, rice caseworm, eat the leaf blade of rice plants and decrease the leaf area. The larvae of leaffolders and rice skippers fold rice leaf blade and remove leaf tissue and make white/transparent streams on the leaf blade, reducing the photosynthetic ability (Pathak and Khan, 1994; Dale, 1994). The adults and nymphs of grasshoppers (Orthoptera), locusts and field crickets can damage leaf blades and, in some circumstances, can cause outbreaks. Other known vegetative pests are rice leaf beetle, whorl maggot, leafminer, thrips and gall midge.
Plant sucking insects: Planthoppers and leafhoppers (Homoptera: Delphacidae and Cicadellidae) are the largest pest group that affect rice cultivation. Nilaparvata lugens, Sogatella furcifera, Laodelphax striatellus and some of the green leafhoppers (Nephotettix species) are distributed across large areas of Asia. Tagosodes orizicolus has been found in the Caribbean islands, South America and the southern United States. They suck the phloem and xylem sap and reduce photosynthesis assimilates in the rice plants. It is well known that infestations of N. lugens cause plant death (hopperburn) when their density on rice plants is extremely high.
The planthoppers and leafhoppers also act as vectors for many viral diseases. For example, N. lugens transmits the grass stunt and ragged stunt viruses in South and Southeast Asia; L. striatellus is a vector for the rice stripe virus and the black-streaked dwarf virus in East Asia; and the green rice leafhoppers are known for being vectors for the tungro viruses in South and Southeast Asia, and rice dwarf virus and yellow dwarf (Phytoplasma) disease in East Asia. S. furcifera is not known as a vector of viruses but, recently, it has been reported that it can transmit Southern rice black-streaked dwarf virus (Zhou et al., 2008; Zhang et al., 2008). Rice black bugs (Scotinophara coarctata, Scotinophara lurida, Hemiptera: Pentatomidae) also feed on plant sap from the rice sheath and reduce plant growth and yield (Joshi, Barrion and Sebastian, 2007).
Grain-sucking insects: After the heading stage of rice plants, many heteropteran insects move to rice paddies from surrounding grassy areas. They usually suck the endosperm of ears of mainly gramineous plants (weeds) around the rice fields. The most important species are rice bugs (Alydidae) and stink bugs (Pentatomidae), as they suck the growing spikelets and cause discolouration of the brown rice which degrades its quality and, in severe cases, sterility of the spikelets. In Japan, the quality degradation of rice grain by the sucking of leaf bugs (Miridae) is also a problem.
Insect pests of upland rice
Soil-inhabiting insects have been recorded in African and Asian countries, such as ants, termites, mole crickets, white grubs (larva of scarab beetle), rice root aphids and rice root weevils. They cause damage when the rice plants are cultivated in upland areas and well-drained conditions (Dale, 1994; Pathak and Khan, 1994).
Host range
The extent of the host range varies greatly among insect species. The planthoppers and the rice stem borers, which are important pests in a wide range of areas, are mostly monophagous or oligophagous. On the other hand, many rice bugs and stink bugs inhabit various gramineous plants and fly to paddy fields during rice heading. The southern green stink bug, Nezara viridula, utilises plants from 32 families and 145 species. In addition, the small brown planthopper, Laodelphax striatellus, lives in gramineous weeds, wheat and rice plants, and it changes hosts depending on the season. Although it is difficult to investigate the actual situation of host plant utilisation in the field, it is important to consider the developmental dynamics of insect species due to the spatiotemporal changes of rice cultivation.
Geographic/genetic variation
The introduction of pest-resistant varieties and the continuous use of pesticides leads to the development of pests that can infest these resistant varieties and are resistant to agricultural chemicals. Regional differences in chemical utilisation also lead to increased genetic variation in pests.
Long-distance migratory species
Some of the rice insect species are known to migrate exceptionally long distances. The brown planthopper N. lugens, whitebacked planthopper S. furcifera, leaffolder Cnaphalocrocis medinalis and armyworm Mythimna separata are representative of long-range migratory pests. N. lugens and S. furcifera pass the winter in the northern part of Viet Nam. After the beginning of the rice cultivation period, they increase their numbers and then start to migrate north to the Korean Peninsula and Japan via the continent. It is reported that rice planthoppers flying to Japan have changed their resistance to pesticides and biotype properties against resistant varieties (Tanaka and Matsumura, 2000; Matsumura et al., 2008; Matsumura and Sanada-Morimura, 2010), which may reflect the history of pesticide usage and resistant varieties in their original source areas. Two bugs, Cyrtorhinus lividipennis Reuter and Tytthus chinensis (Stål), are known to be the major predators of the rice planthoppers in Japan (Nakamura, 2003).
Invasive species
Some pest species are intentionally or unintentionally introduced into new environments from their origins, and cause outbreaks. The rice water weevil Lissorhoptrus oryzophilus, which is native to southern and eastern parts of the United States, was first detected in Aichi Prefecture, Japan, in 1976 and spread rapidly throughout Japan by 1986. It is estimated that they intruded in dry grass and were imported from the United States. It also invaded China and South Korea in 1988. It is now widely distributed in many countries including Chinese Taipei, Greece, India and Italy (Aghaee and Godfrey, 2014; CABI, 2020).
The golden apple snail Pomacea canaliculata (Gastropoda: Ampullariidae) is a large freshwater snail native to South America. After it was introduced to Asian countries for food purposes, individuals escaped and their populations have increased and spread through irrigation systems. It eats young rice plants and destroys the whole plant in the paddy field (Joshi and Sebastian, 2006). Damage to the rice plant is a serious problem in many countries (CABI, 2020) and P. canaliculata has been designated one of the top 100 of the World’s Worst Invasive Alien Species (Invasive Species Specialists Group, 2020).
Predator insects including pollinator and pollen eater
Rice does not have an entomophilous flower but the pollen it produces is used by many organisms. Ladybird beetles and lacewings are natural predators that usually prey on aphids but they also feed on rice pollen when their food supply is low (Pathak and Khan, 1994). A survey in China showed that many insects use rice pollen and that leafcutter bees, sweet bees and honeybees, in particular, carry rice pollen, and Apis mellifera carries pollen for over 500 m (Pu et al., 2014).
Interaction with plants
Weeds
Wild, weedy and volunteer rice plants are described in the first five sections to some extent, from the viewpoints of classification, biology, genetics, introgression and so forth. Therefore, the description related to these plants here focuses on the ecology of wild rice, weedy rice (Oryza sativa L. [f. spontanea] in this document) and volunteer rice (Oryza sativa L. in this document). Weedy and volunteer rice behave as ecological competitors against wild rice in natural ecosystems and weedy rice results in more adverse effects than volunteer rice, from the viewpoint of weediness, such as seed shattering and dormancy. Furthermore, wild, weedy and volunteer rice are the competitors to cultivated rice in the farmland. They voluntarily grow in direct-seeded and transplanted rice areas, although they compete against cultivated rice and are more difficult to manage in direct-seeded rice than transplanted rice, due to the simultaneous growth of the cultivated rice.
Asian O. rufipogon is an ancestor of O. sativa and several factors have contributed to the so-called domestication of rice over a long historic period (Kovach, Sweeney and McCouch, 2007), as explained in the first section.
Throwback (off-type, transmogrify, de-domestication, voluntary) is the opposite of gene flow. It means that the unintentional outcrossing of the cultivated rice with wild relatives resulted in the degradation (off-type) of domestication syndrome towards weedy rice. Several examples have been reported on genetic erosion from cultivated (indica and japonica) cultivars to wild and weedy rice (Suh, Sato and Morishima, 1997; Tang and Morishima, 1998; Ishikawa et al., 2006). Many cross-hybridisations happened to generate weedy type rice or introduce various genetic components (Li et al., 2012; Huang et al., 2012). Some of them have been found as nuclear and cytoplasm substituted lines (Ishikawa et al., 2002a, 2002b; Kim et al., 2015), which may be partly due to the past cross-hybridisation presumed by Li et al. (2017). The introduction of modern varieties into different geographical areas have also resulted in weedy rice (Kawasaki et al., 2009), due to the indica-japonica hybridisation. The hybridisation broke seed shattering because of the inconsistency of gene components.
In addition, weedy rice is geographically distributed in almost all rice-growing areas such as in Brazil, Cambodia, China, Hungary, India, Italy, Japan, Korea, the Lao People’s Democratic Republic, Malaysia, the Philippines, Sri Lanka, Thailand, the United States and Viet Nam, under different cultivation systems including upland/lowland, transplanted/wet sown/dry sown and so forth (Kraehmer et al., 2016). Herbicide-resistant red rice (O. sativa var. sylvatica) against acetolactate synthase (ALS) inhibitors distribute in several countries where Clearfield® rice had been cultivated several years continuously in combination with imazapic and/or imazethapyr (Burgos et al., 2014).
Since rice is grown in a wide range of farmland conditions, such as paddy (shallow/deep water) to upland fields, with different cultivation methods, and wet-/dry-sown to transplanted rice. The favourable conditions are different among the cultivars for the climates in the cultivation areas. Therefore, many kinds of weeds are grown with their own favourite habitats and/or cultivation conditions. Major weeds in the rice fields of the world are listed in Annex 4.C, except for weedy rice (Akanksha, 2009; Caton et al., 2010; IRRI, n.d.-a; n.d.-b; Kraehmer et al., 2016; Moody, 1989; Rao, Chandrasena and Matsumoto, 2017; IRRI, 1983).
The majority of weeds are grasses of Poaceae (Gramineae) species, such as Echinochloa colona, E. crus‑galli, E. glabrescens, Eleusine indica, Ischaemum rugosum, Leptochloa chinensis, Paspalum distichum, followed by sedges (Cyperaceae), such as Cyperus difformis, C. iria, C. rotundus, Fimbristylis miliacea, the other monocotyledons (monocots) and dicotyledons (dicots). In the other monocots, Monochoria vaginalis is the major weed in paddy fields.
Parasitic plants (i.e. Striga spp.) are noxious weeds only under upland conditions, and a diversity of rice genotypes exists in Striga resistance (Gurney et al., 2006; Rodenburg et al., 2017). Aquatic plants, such as algae and floating plants, are also troublesome under shallow water conditions, such as transplanted rice in irrigated paddy conditions.
As these weedy plants are divergent from the Oryza genus, there is no possibility to cross-hybridise. Therefore, the decisive factors of their population dynamics in certain areas are as follows:
Competition between cultivated rice and weeds: Weeds compete with cultivated rice for light, nutrition and water (upland soil condition only). In addition, allelopathy is also one of the important factors (see following sub-section on allelopathic interaction). On the other hand, water depth affects the population dynamics and deep water gives an advantage to cultivated rice growth in general.
Dormancy and longevity of seed and vegetative organs: The dormancy changes under different seasonal, field, buried seed conditions and the longest period was ten years or more depending on the species and the above conditions. The crop rotations between paddy and upland field conditions are also effective tools for changing weed populations, which reduce their longevity and/or the amount of buried seed and vegetative organs.
Mitigation/invasion, acclimation, adaptation ability of weeds: After/at seed shattering, weed seeds are carried by the wind, animals, cultivated soil, rivers and so forth. Cultivated soil can be contaminated with weed seeds attached to the tires of tractors or combine harvesters that move from field to field. Floating seeds in the paddy field flow the outlet to the river via the canal and go downstream. Riverside weeds directly go to the river and take root in the other riverbed in the downstream area. Floating seeds move to the other river basin via the irrigation canal. Import/export is also a crucial route for invasive alien species.
Competitiveness for rice and dormancy are distinctive factors in weeds in comparison to insects and diseases. The adverse effects of weeds on cultivated crops are not dramatic in the short term within the cultivation period but are long term once a weed has established its population in the area.
Allelopathic interaction
There are many reports on the allelopathy of rice. The allelopathic potential of rice might play an important role in improving weed control. There are two ways to research rice allelopathy. One is the screening of allelopathic rice cultivars or accessions. The other is the isolation and identification of allelochemicals from rice plants.
Historically, screening of rice cultivars for their allelopathic potential started in Japan and the United States around 1990. The USDA scientists, Dilday, Nastasi and Smith (1989), Dilday, Mattice and Moldenhauer (2000), Dilday, Lin and Yan (1994) and Dilday et al. (1992, 2001) evaluated allelopathic potential among thousands of rice cultivars collected worldwide. Of these, 412 among 12 000 rice cultivars exhibited allelopathic activity against ducksalad in a field assessment. The strongest cultivars were PI321777 and PI338046. In Japan, Fujii et al. screened allelopathic activity of 500 cultivars of rice in the Gene Bank of Japan by using a bioassay entitled the plant box method and found that traditional red rice such as Awa‑akamai, Kouketsumochi and tropical japonica and African rice (O. glaberrima) possessed greater allelopathic potential to certain weeds than other types, especially the improved types (Fujii, 1992, 2001; Fujii and Shibuya, 1991; Fujii et al., 2001). At IRRI, Olofsdotter et al. tested actual allelopathic activity on the field and found Kouketsumochi showed the strongest suppression activity to certain weeds (Olofsdotter, Navarez and Moody, 1995; Olofsdotter, Navarez and Rebulanan, 1997; Olofsdotter et al., 1999).
Allelopathic potential may be a polygenic characteristic and its correlation with other rice characteristics has been controversial (Dilday et al., 1991). Ebana and Okuno reported QTL analysis with allelopathic rice (Ebana et al., 2001, Okuno and Ebana, 2003). Gu and Guo also did a screening of allelopathic rice varieties (Gu, Wang and Kong, 2008, 2009; Guo et al., 2009). Improved rice cultivars often exert weak allelopathic potential, which may be because of a lack of selection pressure for allelopathic characteristics during breeding (Olofsdotter, Navarez and Moody, 1995).
As for allelochemicals in rice, momilactones were identified from rice straws and leaves (Kato et al., 1973), rice hulls (Cartwright et al., 1981; Chung, Hahn and Ahmad, 2005) and root exudates (Kato-Noguchi and Ino, 2003, 2005; Kato-Noguchi and Peters, 2013). There are many phenolic acids reported. For example, benzoic acid, caffeic acid, salicylic acid and other phenolic acids were found in rice straws (Kuwatsuka and Shindo, 1973). Ferulic acid, coumaric acid, p-hydroxybenzoic acid and salicylic acid were identified in leaves and stems (Chou, Chang and Oka, 1991). Olofsdotter et al. (2002) doubted phenolic compounds as primary allelochemicals in rice because of their low concentration. Bioactive steroids were also reported (Macías et al., 2006). Other many compounds were reported (reviewed in Khanh, Xuan and Chung, 2007; Jabran, 2017; Fujii and Hiradate, 2007). There are many reports on the allelochemical candidates but the contribution of these chemicals was not well examined. There are several papers on how to evaluate the contribution by their total activity defined by the activity of each candidate and concentration in situ (Hiradate, 2006; Fujii and Hiradate, 2005; Hiradate et al., 2010).
Interaction with micro-organisms
Rhizosphere: The rhizosphere soil of rice enables the coexistence of aerobic and anaerobic microbes because the development of root aerenchyma allows microenvironments of aerobic areas in the anaerobic conditions of the paddy field (Shabuer et al., 2015). This also allows for radial O2 loss which makes the difference in the redox potential and has an effect on the biogeochemistry of mineral elements in the rhizosphere, especially for C, N, P, and Fe (Kögel-Knabner et al., 2010).
Methanogens and methanotrophs are important microbes regulating methane dynamics in the rhizosphere. Methanogens that consist of domain archaea, Methanosaeta, Methanocella and Methanobacterium, are the main components (Imchen et al., 2019). It is known that large parts of produced methane are oxidised in the rhizosphere by methanotrophs (Kögel-Knabner et al., 2010). Methylocystis belonging to Type II methanotrophs are reported most abundantly in India (Pandit et al., 2016) and China (Liu et al., 2017b). Many types of nitrogen-fixing microbes have been discovered (59 genera) (Wang et al. 2019) and some of them are considered to play a beneficial role in the growth of rice (Banik, Mukhopadhaya and Dangar, 2016).
Continuous environmental changes from the root-rhizosphere-bulk soil make a large variety of biogeochemical pathways in this region. Nitrification occurred in aerobic conditions by ammonia oxidising archaea (AOA) and/or ammonia oxidising bacteria (AOB). Among AOA, Nitrosocaldus (Imchen et al., 2019) and/or Nitrososphaera (Chen et al., 2008) were reported to be abundant. In the case of AOB, Nitrospira was the most abundant in Japan (Bowatte et al., 2006) and China (Chen et al., 2008). The produced NO3- diffused into adjacent anaerobic conditions, and then denitrifiers led to gaseous nitrogen loss (N2, NO, and N2O) which is strongly dependent on carbon availability (Chen et al., 2018).
Anaerobic ammonia oxidation coupled with Fe3+ reduction, called Feammox, is driven by the Fe3+ reduction (Zhou et al., 2016) and then generates NO2-, NO3- and N2 as the terminal product of the NH3 oxidation pathway using different microbes (Yang, Weber and Silver, 2012).
The existence of fungi is limited under anaerobic conditions, while contributions of arbuscular mycorrhizal fungi (AMF) on the rice growth have been reported (Watanarojanaporn et al., 2013) but it has also been reported that only a few mycorrhizal species were functional under flooded conditions (Gutjahr, Casieri and Paszkowski, 2009). The AMF colonisation rate in the flooded conditions was about one‑third to half that in the non-flooded conditions (Hajiboland, Aliasgharzad and Barzeghar, 2009). The role of the AMF for the rice was not only to increase nutrient uptake but some abiotic and biotic stress was alleviated by the infection (Mbodj et al., 2018). From the metagenomic analysis of the 16s rRNA gene, there was a substantial difference in the composition of rhizosphere micro-organisms within wild rice species and cultivated varieties (Shenton et al. 2016) and, furthermore, when comparing indica (68) and japonica (27) varieties, it was found that nitrogen utilising efficiency was more active under indica varieties (Zhang et al. 2019).
Phyllosphere (Surface area of plant shoot): As the phyllosphere is directly affected by environmental conditions, those microbes in the phyllosphere may act to alleviate the impacts from outside (Vacher et al., 2016). From the phyllosphere, researchers are investigating the beneficial microbes against pathogens (Harsonowati, Astuti and Wahyudi, 2017).
Endophytes: It is important to evaluate the role of endophytes regardless of their cultivability and to address this a metagenomic approach has been carried out to identify the endophytic bacteria of the rice roots (Sessitsch et al., 2015). Based on the metagenomic approach, Gammaproteobacteria, mostly Enterobacter-related endophytes and Alphaproteobacteria, which includes a large number of rhizobia, were identified as the most abundant group. This allowed for the prediction of traits and metabolic processes such as the nitrogen cycle involving nitrogen fixation, denitrification and nitrification.
The carbon cycle was also highlighted: though the relative abundance of methanogens (Methanocella, Methanosarcina and Methanosaeta) was higher in the rhizosphere than inside the rice roots, Methanobacterium was equal or higher inside the root (Edwards et al., 2015).
Influences of rice on organisms in usual close contact
Influences on pathogens
Fungi and oomycetes
Fungal and oomycete diseases (see Annex 4.B)
Rice blast is the most important air-borne paddy rice disease in the world, including in Southeast Asia and the United States. The pathogen is Pyricularia oryzae (syn.: Magnaporthe oryzae), an ascomycete fungus, and infected seeds are the primary infection source; it occurs during the seedling stage. In the rice field, it forms spindle-shaped leaf blast lesions, spores and infects the upper leaves. Infection at the panicle emergence stage causes wilting and death of grain and neck, resulting in a large decrease in yield.
Rice sheath blight is the second most important soil-borne disease in the world, following rice blast. The pathogen is Rhizoctonia solani, a basidiomycete fungus and the sclerotium, which is the primary infection source, floats during pudding and attaches to the rice stem, forming a lesion. The lesions develop on the upper leaves and spread to neighbouring plants. If the disease is severe, even the flag leaves and panicles are affected. This disease can also cause lodging in strong winds such as typhoons.
Rice false smut is a disease that occurs in China and Southeast Asia and forms black spore balls on rice grains during the ripening stage. The pathogen is Ustilaginoidea virens, which belongs to Ascomycota. Chlamydospores contained in diseased grains fall into the soil and become the primary infection source. When rice plants are transplanted the following year, the fungus invades through the roots and reaches the spikelets during the panicle formation stage, leading to the disease.
Rice brown spot is an air-borne disease that occurs mainly in South and Southeast Asia. The pathogen is Cochliobolus miyabeanus, which belongs to Ascomycota. The primary infection source is infected seed and diseased straw. The fungus causes brown spots on leaves and when panicles are infected, panicles may die. The disease occurs often in soils that are deficient in microelements and fertilisers (Ou, 1985; Cartwright et al., 2018).
Mechanisms of symptom development and rice resistance for blast disease
The causal agent of blast disease, Pyricularia oryzae (syn.: Magnaporthe oryzae), is an ascomycete whose genome has been sequenced and made available to the public. The host-pathogen interactions in this disease are controlled under “Gene-for-Gene” interactions. Rice is resistant (incompatible interaction) or susceptible (compatible interaction) to P. oryzae if rice has a true resistance gene (R-gene, Kalia and Rathour, 2019) that recognises the fungal race-specific “effector” gene, or not respectively. Among these effectors, AvrPii and AvrPiz-t have been characterised for their biochemical roles in the host cell (Park et al., 2012, Singh et al., 2016). Thus, resistance regulated by the R‑gene, called effector-triggered immunity (ETI), is highly race-specific. By 2019, 25 R‑genes were cloned and characterised (Kalia and Rathour, 2019). The fungal invasion starts with the formation of dome-shaped specific structures, the appressorium, from which infectious hyphae penetrate the host epidermal cell (Howard and Valent, 1996).
In the incompatible interaction, hyphal extension is strongly restricted at the early stage of infection by programmed death of invaded rice cells, while in the compatible interaction, the fungal hyphae penetrate into the rice cell, keeping the plasma membrane and organelle, including the vacuole, intact. This observation strongly indicates that the virulent race of this fungus is able to suppress the host defence (Yan and Talbot, 2016).
Another class of immunity is called PAMP-triggered immunity (PTI), where cell components of pathogens, PAMPs (Pathogen-Associated Molecular Patterns), induce defence responses in rice (Liu et al., 2013). PTI is not race-specific and a main part of basal resistance. The most studied PAMP from P. oryzae is the chitin oligomer, hydrolysate of chitin which is a backbone structure of fungal cell walls. Rice recognises this elicitor using two sensors, OsCEBiP and OsCERK1 (Desaki et al., 2018). Recognition of chitin oligomers by rice have been demonstrated to contribute to basal resistance against P. oryzae (Kishimoto et al., 2010).
On the other hand, P. oryzae has developed novel strategies to avoid the host resistance induced by chitin oligomers. One is the masking of cell wall chitin with α‑1,3‑glucans after starting hyphal penetration into the host cell. As higher plants do not have α‑1,3‑glucanase activities, P. oryzae can protect the chitin backbone in the cell wall from attack by chitinase of the rice origin leading to the production of chitin oligomers (Fujikawa et al., 2012). Another strategy of the fungus is Slp1 secreted from infectious hyphae. This protein has a high binding affinity to chitin oligomers and is considered to contribute to successful infections by preventing chitin oligomers from being recognised by CEBiP (Mentlak et al., 2012).
Both ETI and PTI of higher plants induce largely common defence responses such as the expressions of pathogenesis-related (PR) protein genes and the accumulation of anti-microbial metabolites, phytoalexins (Peng, van Wersch and Zhang, 2018). Therefore, the signalling pathway likely merges into a common pathway after perception of the effectors or PAMPs by R-gene or receptors respectively. Several key factors in the common signal pathway have been identified. A small GTP-binding protein, OsRac1, has been demonstrated to be deeply involved in both ETI and PTI through its activation/inactivation cycle (Liu et al., 2013). Salicylic acid (SA) was also observed to play essential roles in the defence responses in rice. Transgenic rice harbouring the SA-inactivating gene exhibits compromised resistance in ETI and PTI against P. oryzae. In the downstream of SA signalling, two signalling factors, OsNPR1 and OsWRKY45, have been identified as key factors. Constitutive expression of OsWRKY45 confers strong resistance to the infection of Xanthomonas oryzae, a bacterial pathogen of rice leaf blight disease, in addition to P. oryzae (Takatsuji, H., 2014).
Bacterial pathogen (see Annex 4.B)
Eleven species of bacteria have been identified as a pathogen of rice and the site of infection for all bacteria is the above-ground parts of the plant. The main bacterial diseases of rice are bacterial grain rot, bacterial brown stripe, bacterial seedling blight, bacterial blight (BB) and bacterial leaf streak.
The bacterial grain rot is caused by Burkholderia glumae and occurs at the seedling stage. The leaf sheaths or leaf blades turn light or dark brown and decompose. Alternatively, the leaves become yellowing and curling, eventually leading to death. Symptoms initially appear in patches but then spread to the surrounding area. After the ear emergence stage (after the milk-ripening stage), the disease symptoms also appear on the panicle. The panicle wilts to a white, greyish-white or light yellowish-brown colour, resulting in poor fertility. Burkholderia glumae is a gram-negative, rod-shaped aerobic bacterium with an optimal growth temperature of around 30°C and an optimal pH of 6.0-7.5. Under natural conditions, rice is the only host of this bacterium, which is transmitted from infested seeds or soil and infects the panicle through the leaf sheath and leaf blade (Tsushima, 1996; Ura et al., 2006).
The bacterial brown stripe, caused by Acidovorax avenae, forms brown, elongated, streaky lesions on leaf sheaths and leaf blades of seedlings, resulting in stunting of growth and death. Subsequently, the whole plant turns brown mainly from curled leaf sheaths and brown streaky lesions, leading to death, but the disease is only dispersed throughout nursery boxes. Acidovorax avenae prefers high temperatures (optimal growth temperature: 35-40°C) and is transmitted by seeds (Kadota and Ohuchi, 1990).
The bacterial seedling blight caused by Burkholderia plantarii occurs only during seedling growth in nursery boxes and does not occur in adult rice. The early symptoms are browning of the basal part, chlorosis and wilting at the base of new leaves but no rotting at the base. Subsequently, leaves roll, wilt and turn brown, leading to death. In addition, this disease produces a toxin (tropolone) that inhibits root elongation and above-ground greening and often occurs in spots. This disease is transmitted from infected seeds of the previous year and secondary infection is promoted by high temperatures during germination and emergence. Although the disease develops remarkably at high temperatures (30-34°C) during germination and seedling growth, it develops less severely at temperatures below 30°C and does not develop at temperatures above 37°C. In addition, the optimum pH for this disease is lower than 5.0‑5.5 (Azegami et al., 1987).
The BB is caused by Xanthomonas oryzae pv. oryzae which is a gram-negative, rod-shaped bacterium. The bacteria enter the leaf through the hydathodes or wounds, multiply in the intercellular spaces of the underlying epitheme, and propagate to reach the xylem vessels. They further move through the veins of leaves and spread into the plant. The water-soaked spots at the leaf tips and margins are first observed and then, the leaves become chlorotic and necrotic along the leaf veins. The bacteria can pass the winter in the seed, straw, stubble or the soil, but in case the disease is induced next year, the major origin is the Poaceae (Gramineae) family weeds that are growing in the ridge of the field or the irrigation canal. The bacteria enter the field on the water flow and thereafter are spread by the wind. If the rice nursery can be flooded easily, an excess of nitrogen during the fertilisation process stimulates a rapid vegetative overgrowth of the rice plants that favours the disease development (Niño-Liu Zohary, Ronald and Bogdanove, 2006).
The bacterial leaf streak is caused by Xanthomonas oryzae pv. oryzicola, a gram-negative rod-shaped bacterium with an optimal growth temperature of 25-28°C. The bacteria penetrate the leaf mainly through stomata or wounds, multiply in the substomatal cavity and then colonise the intercellular spaces of the parenchyma. Different from the BB, small, water-soaked lesions along the leaf between the veins were observed during the early stage of bacterial leaf streak infection, resulting in translucent and yellow streaks. The infected leaves turn greyish-white and die later on. This disease frequently occurs in the condition of high temperature and humidity, and, in severe cases, the field turns brown entirely (Niño-Liu, Ronald and Bogdanove, 2006).
Phytoplasmas (see Annex 4.B)
Two species of phytoplasmas have been identified as pathogens of rice, one for Candidatus Phytoplasma oryzae causing yellow dwarf and the other for Candidatus Phytoplasma asteris causing orange leaf.
The rice yellow dwarf phytoplasma is mediated by green rice leafhopper, for which the transovarial transmission does not occur. The symptoms of the disease are characterised by prominent stunting of plants and excessive tillering. Leaf colour changes from yellowish green to whitish green, and the leaf becomes soft and droops. The disease is transmitted by leafhopper vectors Nephotettix sp. with a latent period of 25‑30 days in the vector. The pathogen also survives on several grass weeds (Muniyappa and Raychaudhuri, 1988; Nakashima and Hayashi, 1995).
Rice orange leaf disease phytoplasma causes moderate stunting and the appearance of a golden or orange leaf colouration that initiates at the tip and then progresses downward, followed by an inward rolling of leaves and eventually leading to leaf senescence in mature rice plants. Then the grain yield is seriously damaged. This disease also occurs at the seedling stage and often causes a lethal effect. Insects such as zigzag-striped leafhopper (Recilia dorsalis Motschulsky) and green leafhopper (Nephotettix cincticeps Uhler) are responsible for the spread of this phytoplasma (Valarmathi et al., 2013; Jonson et al., 2020).
Viruses (see Annex 4.B)
There are approximately 14 species of viruses that infect rice. The major vectors are pests, while in some cases, can be transmitted transovarially to their offspring. The characteristics of the major viruses are described below:
The Rice dwarf virus (RDV) is transmitted among rice plants by green rice leafhopper (Nephotettix cincticeps). The virus multiplies in the pests and can be transmitted transovarially to their offspring (Honda et al., 2007). The virus infection occurs mainly just after the transplanting, through the feedings damage caused by infected leafhoppers. The infected rice plants transmit the virus to other rice plants via the pests, and the pests passing the winter transmit the virus to rice plants the following year. The symptom is the dwarfing of the stubble at the tillering stage, change in leaf colour to dark green and display of a series of vivid white spots along the leaf veins. In addition, if the infection occurs at the early stage of rice growth, the rice does not head, or even if the heading occurs, the panicle is small and becomes sterile (Morales, 2008).
Rice ragged stunt virus (RRSV) is a double-stranded RNA virus that is classified in the Oryzavirus genus. It is transmitted by Nilaparvata lugens and transovarial transmission does not occur. The disease occurs in China, Chinese Taipei, India, Indonesia, Japan, Malaysia, Thailand and the Philippines. The infection causes dwarfing of the whole stubble, change in leaf colour to dark green, and serration and twisting of the leaves (Hibino et al., 1986a).
The Rice black-streaked dwarf virus (RBSDV) is a double-stranded RNA virus that is classified in the Fijivirus genus. It is transmitted by Laodelphax striatellus and causes damage in China, Japan and Korea. This virus also causes damage to the corn. The infection induces extreme stunting, darkening of the leaves and twisting of the distal portions of young rice leaves (Wu et al., 2020). During the last few years, Southern rice black-streaked dwarf virus (SRBSDV), a species that is closely related to the RBSDV and mediated by Sogatella furcifera, has rapidly spread throughout China, Japan and Viet Nam (Zhou et al., 2013).
The Rice stripe virus (RSV) is an RNA virus that is classified in the Tenuivirus genus. This virus is spreading throughout East Asia, especially in China, Japan and Korea. The virus is mediated mainly by small brown planthopper Laodelphax striatellus or other planthoppers such as Unkanodes sapporona or Terthron albovittatum. The transmission occurs transovarially but neither seed transmission nor contagious transmission occurs. The major hosts of this virus are crops and weeds of the Poaceae (Gramineae) family. The disease frequently occurs at the tillering stage and the RSV-infected plants display chlorosis, weakness and necrosis in leaves, abnormal growth and result in death (Cho et al., 2013).
Rice grassy stunt virus (RGSV) is an RNA virus that is classified in the Tenuivirus genus. The disease is occurring widely in East and Southeast Asia, including China, India, Japan and Sri Lanka. The virus is transmitted by Nilaparvata lugens and the transovarial transmission does not occur. The infection causes yellowing of the leaf, dwarfing of the plant, browning/dark-browning and poor fertility of the panicle (Hibino, 1986b).
Rice tungro disease is causing damage in South and Southeast Asia, including Bangladesh and India. This disease is mediated by Nephotettix impicticeps and results in the yellowing of the leaf and leaf sheaths and dwarfing of the whole stubble. The disease is caused by the combination of two viruses, one for Rice tungro bacilliform virus (RTBV), classified in the Tungro genus and involved in pathogenesis, and the other for Rice tungro spherical virus (RTSV), classified in the Waikavirus genus and involved in the virus transmission (Hibino, 1983).
Influences on invertebrate pests
Plants have evolved a range of defence mechanisms to protect themselves from damage by herbivores (Mithöfer, Boland and Maffei, 2009; Erb and Reymond, 2019). Two such mechanisms of defence are mechanical protection and chemical protection. Plant defences can be further categorised into constitutive and induced defences following herbivore feeding (Mithöfer, Boland and Maffei, 2009). Insect-resistant varieties of rice have been screened to determine which varieties have resistance to which insect pests. It has been clarified that there are many varieties resistant to specific insect pests and that this is variety dependent (Heinrichs, Medrano and Rapusas, 1985).
Many species of sap-sucking pest insects ingest nutrients from the phloem of rice. Some rice varieties are resistant to these insect pests, for example delphacid planthoppers (the brown planthopper [BPH] N. lugens, the whitebacked planthopper [WBPH] S. furcifera and the small brown planthopper [SBPH] L. striatellus) and cicadellid leafhoppers (the green rice leafhopper [GRH] Nephotettix cincticeps and the green leafhopper [GLH] Nephotettix apicalis(. Some of the genes involved in rice resistance to insect pests have been mapped and used for breeding (Fujita, Kohli and Horgan, 2013). Secondary metabolites thought to be related to plant constitutive defence mechanisms have been analysed to reveal causative factors of varietal resistance. However, different sources reported that it is induced resistance that contributes to varietal resistance (Kaloshian and Walling, 2016; Ling, Ang and Weilin, 2019; Du et al., 2020). In this subsection, we focus mainly on BPH and GRH, and introduce studies on the resistance response of rice to these insect pest species.
Since the 1960s, many BPH resistant rice varieties have been discovered (Pathak and Khush, 1979). Based on the hypothesis that resistant rice varieties must contain feeding deterrents to pest species, it was first attempted to isolate feeding deterrents from resistant varieties using natural product chemistry techniques. Following these lines of enquiry, Yoshihara et al. (1980) reported oxalic acid as a feeding deterrent in rice leaf sheaths that carried the BPH resistance gene, Bph1. In addition, Shigematsu et al. (1982) reported β-sitosterol as a further feeding deterrent. Later, Stevenson et al. (1996) identified schaftoside as a feeding deterrent from the indica rice variety Rathu Heenati that carries the resistance gene Bph3. These feeding deterrent compounds are important substances in constitutive defences of rice against herbivorous insects. If these feeding deterrents were responsible for varietal resistance, then we would expect that the virulent biotype of BPH, which is known to feed on resistant varieties, would be adapted to these feeding deterrents. However, it has not yet been confirmed that these deterrents are not effective against the virulent biotype.
The feeding behaviour of BPH and GRH on resistant varieties has been analysed in detail using electric penetration graphs (Kawabe, 1985; Hattori, 2001). BPH is a monophagous insect that only feeds on rice. When BPH attempts to attack a non-host plant, such as the barnyard grass (Echinochloa crus-galli var. oryzicola), BPH probing is interrupted before the arrival of the stylets at the sieve elements of the rice (Hattori, 2001). It is thought that probing is interrupted by a feeding deterrent such as (E)-aconitic acid in the parenchyma of rice (Hattori, 2001). On the other hand, although the stylet mouth part of BPH reaches the sieve elements of the resistant rice variety, BPH can hardly suck the phloem sap of the resistant rice. Therefore, this sucking inhibition by resistant rice likely does not occur in the parenchyma but in the phloem sieve elements. Hattori (1997) reported that GRH could suck all of the phloem sap that was collected from three different GRH-resistant rice varieties by a stylectomy method using BPH that can feed on the GRH-resistant variety. Thus, it seems that non-constituents that are particularly unsuitable as gustatory stimuli are involved in the phloem sap of the tested GRH-resistant varieties.
Owing to genomic information, some BPH resistance genes have been isolated and characterised from BPH resistant rice varieties and wild rice spices (Ling et al., 2019; Du et al., 2020). Bph14 derived from wild rice, which was first isolated as a resistance gene for BPH, and BPH26, which was subsequently isolated from an indica rice variety, encode nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins (Du et al., 2009; Tamura et al., 2014).
Most of the disease resistance genes in plants encode NBS-LRR proteins and they are considered to induce a host plant defence response through directly or indirectly recognising a pathogen-derived effector (Elmore, Lin and Coaker, 2011). Since BPH resistance proteins are also NBS-LRRs, it is expected that they recognise injury signals, such as effectors, from BPH and induce the defence response, the same as for the disease resistance proteins. It is suggested that ETI may be present in the defence of rice against sucking insects as NBS-LRRs have been isolated as resistance proteins from rice.
When fed on rice, BPH secretes two kinds of saliva: gelling and watery saliva (Huang et al., 2016). The BPH resistance protein of the NBS-LRR family is predicted to recognise a saliva protein as an effector and become active. As a result, it induces sucking inhibition in the sieve elements of rice plants.
The BPH14 protein is thought to form homocomplexes that interact with the transcription factors WRKY46 and WRKY72 (Hu et al., 2017). WRKY46 and WRKY72 then bind to the promoters of the receptor-like cytoplasmic kinase gene RLCK281 and the callose synthase gene LOC_Os01g67364.1, whose transactivation activity is dependent on WRKY46 or WRKY72. Sieve element occlusion through callose deposition is thought to be an important defence mechanism, induced by Bph14, which prevents planthoppers from ingesting phloem sap (Du et al., 2009; Hu et al., 2017).
The defence response mediated by the NBS-LRR resistance protein is characterised by a rapid, high specificity response and strong resistance. In resistant varieties carrying the NBS-LRR resistance protein, BPH does not settle and, when BPH is forcibly attached, the mortality rate of BPH increases, egg production decreases and the rice plant survives. These phenomena observed in resistant varieties are caused by an inhibition of sucking in the phloem (Sōgawa, 1982). It is expected that BPH survival and the development of the ovaries is affected by nutritional deficiency.
Other non-NBS-LRR BPH resistance genes have been cloned, suggesting that there may be various forms of plant defence responses other than ETI. Bph3 (originally reported as Bph17), which was cloned from the indica rice variety Rathu Heenati, is a plasma membrane-localised lectin receptor kinase (OsLecRK1-OsLecRK3; Liu et al., 2015). BPH15 is also thought to be a lectin receptor kinase (Du et al., 2020). Bph3 is characterised by broad-spectrum and durable insect resistance. Although it is unknown what the ligands of lectin receptor kinases are, they may play a critical role in priming plant pattern-triggered immunity (PTI) responses to BPH infestation (Liu et al., 2015).
BPH6 encodes a previously uncharacterised protein that localises to exocysts and interacts with the exocyst subunit OsEXO70E1 (Guo et al., 2018). BPH6 expression facilitates exocytosis and cell wall reinforcement and induces co‑ordinated salicylic acid, cytokinin and jasmonic acid signalling. This gene is effective not only for BPH but also for WBPH. BPH29 and Bph32 also encode proteins different from NBS-LRR (Wang et al., 2015; Ren et al., 2016). Lu et al. (2018) have also reported that the resistance of rice to insect pests may be mediated by the suppression of serotonin biosynthesis.
Rice likely has various defence pathways against sucking insects and these have coevolved between rice and sucking insects over many years. Although there are still many unclear points concerning rice defence mechanisms to insect pests, the culmination of these research studies will likely lead to the production of rice varieties that are insect pest resistant.
Annex 4.A. Glossary of rice ecological types and their relationships
Annex Table 4.A.1. Characteristics of rice ecological types
|
Ecological type of rice |
Description |
Habitat |
|
|---|---|---|---|
|
Natural ecosystem |
Farmland |
||
|
Wild rice |
Ancestor and close relatives of cultivated rice which hold seed shattering and dormancy. |
+++ |
+ |
|
Weedy rice |
Mixture of wild rice and hybrid offspring derived from wild x wild, wild x cultivar and cultivar x cultivar. They are difficult to distinguish by species classification. Some weedy rices are cultivated rice but lost domestication characteristics due to outcrossing with wild relatives and others are adapted wild rice to the farmland. |
+ |
+++ |
|
Volunteer rice |
Voluntarily emerged cultivated rice. They are derived from dropped and buried seeds in the previous years, because of their seed shattering and dormant activities. |
+ |
+++ |
|
Cultivated rice |
Cultivars domesticated from wild rice. Their traits are almost fixed in each cultivar under cultivation condition. |
- |
+++ |
Annex Figure 4.A.1. Relationships among wild, weedy, volunteer and cultivated rice
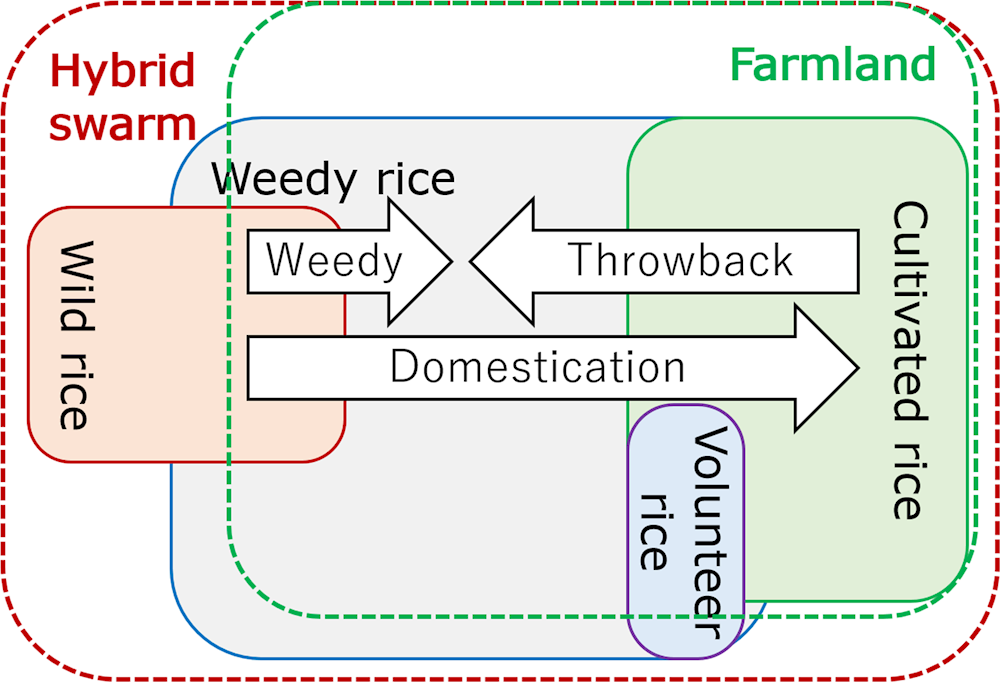
Annex 4.B. Rice diseases
Annex Table 4.B.1. Fungal and oomycete diseases
|
Common name of disease |
Scientific name of pathogen |
Affected parts |
|---|---|---|
|
Aggregate sheath spot (Brown sclerotial disease) |
Ceratobasidium setariae (Sawada) Oniki et al. (syn. Ceratobasidium oryzae-sativae P. S. Gunnell & R. K. Webster) |
Sheath, Culm |
|
Bakanae |
Fusarium fujikuroi Nirenberg (syn. Gibberella fujikuroi (Sawada) S. Ito) |
Seedling, Crown, Lower culm |
|
Black kernel (Brown blotch of grains) |
Curvularia clavata B.L. Jain Curvularia inaequalis (Shear) Boedijn Curvularia intermedia Boedijn Curvularia lunata (Wakker) Boedijn Curvularia ovoidea (Hiroë) Munt.-Cvetk. Alternaria alternata (Fr.) Keissl. |
Grain |
|
Blast |
Pyricularia oryzae Cavara (syn. Magnaporthe oryzae B. C. Couch) |
Seedling, Leaf, Culm, Panicle |
|
Brown spot |
Bipolaris oryzae (Breda de Haan) Shoemaker (syn. Cochliobolus miyabeanus (Ito & Kurib.) Drechsler ex Dastur) (syn. Drechslera oryzae (Breda de Haan) Subram. & P. C. Jain) (syn. Helminthosporium oryzae Breda de Haan) |
Seedling, Leaf, Panicle |
|
Crown sheath rot (Black sheath rot) |
Gaeumannomyces graminis (Sacc.) Arx & D. L. Olivier |
Lower leaf sheath |
|
Downy mildew |
Sclerophthora macrospora (Sacc.) Thirum. et al. (syn. Sclerospora macrospora Sacc.) |
Leaf |
|
Eyespot |
Drechslera gigantea S. Ito |
Leaf |
|
False smut |
Ustilaginoidea virens (Cooke) Takah. (syn. Villosiclava virens (Nakata) E. Tanaka & C. Tanaka) |
Grain |
|
Glume blight |
Microsphaeropsis glumarum (Ellis & Tracy) Boerema Epicoccum sorghinum (Sacc.) Aveskamp et al. |
Grain |
|
Kernel smut |
Tilletia barclayana (Bref.) Sacc. & P. Syd. (syn. Neovossia barclayana Bref.) (syn. Tilletia horrida Takah.) |
Grain |
|
Kernel discolouration (Grain discolouration, black kernel) |
Alternaria padwickii (Ganguly) M.B. Ellis |
Grain |
|
Leaf scald |
Microdochium albescens (Thüm.) Hern.-Restr. & Crous (syn. Gerlachia oryzae (Hashioka & Yokogi) W. Gams) |
Leaf, Sheath |
|
Leaf smut |
Eballistra oryzae (Syd. & P. Syd.) R. Bauer et al. (syn. Entyloma oryzae Syd. & P. Syd.) |
Leaf |
|
Red blotch of grains |
Epicoccum nigrum Link (syn. Epicoccum purpurascens Ehrenb.) Epicoccum neglectum Desm. Epicoccum oryzae S. Ito & Iwadare |
Grain |
|
Red stripe |
Gonatophragmium sp. Some papers attributed the disease to bacteria. |
Leaf |
|
Scab |
Fusarium graminearum Schwabe (syn. Gibberella zeae (Schwein.) Petch) |
Grain, Culm |
|
Seedling blight (Seedling damping-off) |
Fusarium spp. Pythium spp. Globisporangium spinosum Uzuhashi et al. (syn. Pythium spinosum Sawada) Rhizopus spp. Mucor fragilis Bainier Trichoderma viride Pers. Rhizoctonia solani J.G. Kühn Athelia rolfsii (Curzi) C. C. Tu & Kimbr. (syn. Sclerotium rolfsii Sacc.) |
Seedling |
|
Sheath blight |
Rhizoctonia solani J.G. Kühn (syn. Thanatephorus cucumeris (A. B. Frank) Donk) |
Sheath, Leaf |
|
Sheath blotch |
Sydowia polyspora (Bref. & Tavel) E. Müll. |
Sheath |
|
Sheath net blotch |
Calonectria morganii Crous et al. (syn. Cylindrocladium scoparium Morg.) |
Sheath |
|
Sheath rot |
Sarocladium oryzae (Sawada) W. Gams & D. Hawksw. (syn. Acrocylindrium oryzae Sawada) (syn. Sarocladium attenuatum W. Gams & D. Hawksw.) |
Flag leaf sheath, Grain |
|
Sheath spot |
Waitea circinata Warcup & P.H.B. Talbot (syn. Rhizoctonia oryzae Ryker & Gooch) |
Sheath |
|
Stackburn (Alternaria leaf spot) |
Alternaria padwickii (Ganguly) M.B. Ellis |
Leaf |
|
Stem rot |
Nakataea oryzae (Catt.) J. Luo & N. Zhang (syn. Magnaporthe salvinii (Catt.) R. A. Krause & R. K. Webster) |
Stem |
|
Udbatta (Black choke, incense rod, false ergot) |
Balansia oryzae-sativae Hashioka (syn. Balansia oryzae (Syd.) Naras. & Thirum.) |
Panicle |
|
Water moulds |
Achlya spp. Pythium app. Dictyuchus spp. Globisporangium spinosum Uzuhashi et al. Others |
Seed, Seedling |
|
White leaf streak |
Mycovellosiella oryzae (Deighton & D.E. Shaw) Deighton (syn. Ramularia oryzae Deighton & D. E. Shaw) |
Leaf |
Annex Table 4.B.2. Bacterial diseases
|
Common name of disease |
Scientific name of pathogen |
Affected parts |
|---|---|---|
|
Bacterial brown stripe |
Acidovorax avenae subsp. avenae (Manns) Willems et al. (syn. Pseudomonas avenae Manns) |
Seedling |
|
Bacterial foot rot |
Dickeya zeae Samson et al. (syn. Erwinia chrysanthemi pv. zeae (Sabet) Victoria et al. |
Node, Culm, Crown |
|
Bacterial grain rot(Bacterial seedling rot) |
Burkholderia glumae (Kurita & Tabei) Urakami et al. (syn. Pseudomonas glumae Kurita & Tabei) Burkholderia gladioli (Severini) Yabuuchi et al. (syn. Pseudomonas gladioli Severini) |
Spikelet, Seed, Seedling |
|
Bacterial leaf blight |
Xanthomonas oryzae pv. oryzae (Ishiyama) Swings et al. |
Leaf |
|
Bacterial leaf streak |
Xanthomonas oryzae pv. oryzicola (Fang et al.) Swings et al. |
Leaf |
|
Bacterial palea browning |
Pantoea ananatis (Serrano) Mergaert et al. (syn. Erwinia herbicola (Löhnis) Dye) |
Panicle |
|
Bacterial seedling blight |
Burkholderia plantarii (Azegami et al.) Urakami et al. |
Seedling |
|
Halo blight |
Pseudomonas syringae pv. oryzae (Kuwata) Young et al. |
Leaf |
|
Red stripe |
Microbacterium sp. Some papers attributed the disease to fungi. |
Leaf |
|
Sheath brown rot |
Pseudomonas fuscovaginae Miyajima et al. |
Sheath (mainly flag leaf) |
|
Stem necrosis |
Pantoea ananatis Serrano |
Node |
Annex Table 4.B.3. Phytoplasmal diseases
|
Common name of disease |
Scientific name of pathogen |
Vector |
|---|---|---|
|
Orange leaf |
Candidatus Phytoplasma asteris |
Zig-zag leafhopper |
|
Yellow dwarf |
Candidatus Phytoplasma oryzae |
Green rice leafhopper (temperate) Green leafhopper (tropics) |
Annex Table 4.B.4. Viral diseases
|
Common name of disease |
Scientific name of pathogen |
Vector |
|---|---|---|
|
Black-streaked dwarf |
Rice black-streaked dwarf virus genus Fijivirus; family Reoviridae |
Small brown planthopper |
|
Dwarf |
Rice dwarf virus genus Phytoreovirus; family Reoviridae |
Green rice leafhopper Green leafhopper Zig-zag leafhopper |
|
Gall dwarf |
Rice gall dwarf virus genus Phytoreovirus; family Reoviridae |
Green leafhopper Zig-zag leafhopper |
|
Giallume |
Barley yellow dwarf virus genus Luteovirus; family Luteoviridae |
Bird cherry-oat aphid |
|
Grassy stunt |
Rice grassy stunt virus genus Tenuivirus; family Phenuiviridae |
Brown planthopper |
|
Hoja blanca |
Rice hoja blanca virus genus Tenuivirus; family Phenuiviridae |
Rice delphacid |
|
Necrosis mosaic |
Rice necrosis mosaic virus genus Bymovirus; family Potyviridae |
Polymyxa graminis |
|
Ragged stunt |
Rice ragged stunt virus genus Oryzavirus; family Reoviridae |
Brown planthopper |
|
Southern black-streaked dwarf |
Southern rice black-streaked dwarf virus genus Fijivirus; family Reoviridae |
Small brown planthopper Whitebacked planthopper |
|
Stripe |
Rice stripe virus genus Tenuivirus; family Phenuiviridae |
Small brown planthopper |
|
Stripe necrosis |
Rice stripe necrosis virus genus Benyvirus; family Benyviridae |
Polymyxa graminis |
|
Tungro (dual infection) and Waika |
Rice tungro bacilliform virus genus Tungrovirus; family Caulimoviridae Rice tungro spherical virus genus Waikavirus; family Secoviridae |
Green leafhopper Green rice leafhopper Zig-zag leafhopper |
|
Yellow mottle |
Rice yellow mottle virus genus Sobemovirus; family Solemoviridae |
Adult chrysomelid beetles |
|
Yellow stunt (formerly Transitory yellowing) |
Rice yellow stunt nucleorhabdovirus genus Nucleorhabdovirus; family Rhabdoviridae |
Green rice leafhopper Green leafhopper |
Sources (annex): American Phytopathological Society (2017), Diseases of Rice (Oryza and Zizania spp.), Common Names of Plant Diseases, https://www.apsnet.org/edcenter/resources/commonnames/Pages/Rice.aspx (accessed 11 June 2019); Cartwright, R.D. et al. (2018), Compendium of Rice Diseases and Pests, Second edition, APS Press, p. 145; Hiraguri, A. et al. (2010), “Complete sequence analysis of rice transitory yellowing virus and its comparison to rice yellow stunt virus”, https://doi.org/10.1007/s00705-009-0557-8; Index Fungorum (2019), The Global Database of Fungal Names, http://www.indexfungorum.org/ (accessed 11 June 2019); ICTV (2019), International Committee on Taxonomy of Viruses, https://talk.ictvonline.org (accessed 11 June 2019); IRRI (2016), Rice Diseases: Biology and Selected Management Practices, http://rice-diseases.irri.org/home/contents; Xie, L. and J. Lin (1980), “Studies on rice bunchy stunt disease of rice, a new virus disease of rice plant”, https://doi.org/10.1360/sb1980-25-9-785; Zhu, Y. et al. (2017), “Draft genome sequence of rice orange leaf phytoplasma from Guangdong, China”, https://doi.org/10.1128/genomeA.00430-17.
Annex 4.C. Rice pests
Annex Table 4.C.1. Arthropoda
|
Scientific name |
Common name (English) |
|
|---|---|---|
|
Hemiptera (planthoppers, leafhoppers and others) |
||
|
Alydidae |
Leptocorisa acuta Thunberg |
Rice bug |
|
Leptocorisa chinensis Dallas |
||
|
Leptocorisa oratorius Fabricius |
||
|
Stenocoris claviformis Ahmad |
||
|
Stenocoris southwoodi Ahmad |
||
|
Aphididae |
Hysteroneura (=Carolinaia) setariae Thomas |
Rusty plum aphid |
|
Tetraneura nigriabdominalis Sasaki |
Rice root aphid |
|
|
Cicadellidae |
Cicadella viridis Linnaeus |
Green leafhopper |
|
Cofana (=Tettigella=Cicadella) spectra Distant |
White leafhopper |
|
|
Nephotettix cincticeps Uhler |
Green rice leafhopper |
|
|
Nephotettix malayanus Ishihara & Kawase |
Green leafhopper |
|
|
Nephotettix nigropictus (=apicalis) Stål |
Green leafhopper |
|
|
Nephotettix virescens (=impicticeps) Distant |
Green leafhopper |
|
|
Recilia dorsalis Motschulsky |
Zigzag leafhopper |
|
|
Delphacidae |
Laodelphax striatellus Fallén |
|
|
Nilaparvata lugens Stål |
||
|
Sogatella furcifera Horváth |
||
|
Tagosodes (=Sogatodes) orizicolus Muir |
||
|
Miridae |
Stenotus rubrovittatus Matsumura |
Sorghum plant bug |
|
Trigonotylus caelestialium (Kirkaldy) |
Rice leaf bug |
|
|
Pentatomidae |
Eysarcoris (=Stollia) ventralis Westwood |
White-spotted stink bug |
|
Nezara viridula Linnaeus |
Southern green stink bug |
|
|
Oebalus pugnax Fabricius |
Rice stink bug |
|
|
Pygomenida varipennis Westwood |
Stink bug |
|
|
Scotinophara coarctata Fabricius |
Malayan rice black bug |
|
|
Scotinophara lurida Burmeister |
Japanese rice black bug |
|
|
Pseudococcidae |
Brevennia (=Heterococcus=Ripersia) rehi (=oryzae) Lindinger |
Rice mealybug |
|
Pseudococcus saccharicola Takahashi |
Rice mealybug |
|
|
Thysanoptera (thrips) |
||
|
Phlaeothripidae |
Haplothrips aculeatus Fabricius |
Rice aculeated thrips |
|
Thripidae |
Stenchaetothrips (=Baliothrips=Thrips) biformis Bagnall |
Rice thrips |
|
Lepidoptera (moths) |
||
|
Crambidae |
Chilo auricilius Dudgeon |
Gold-fringed stem borer |
|
Chilo polychrysus Meyrick |
Dark-headed striped stem borer |
|
|
Chilo partellus Swinhoe |
Sorghum stem borer |
|
|
Chilo suppressalis Walker |
Striped stem borer |
|
|
Chilo zacconius Bleszynski |
Striped stem borer |
|
|
Cnaphalocrocis medinalis Guenée |
Rice leaffolder |
|
|
Diatraea saccharalis Fabricius |
Sugarcane borer |
|
|
Nymphula depunctalis Guenée |
Rice caseworm |
|
|
Marasmia (=Susumia) exigua Butler |
Rice leaffolder |
|
|
Marasmia patnalis Bradley |
Rice leaffolder |
|
|
Marasmia trapezalis Guenée |
Rice leaffolder |
|
|
Parapoynx diminutalis Snellen |
Rice caseworm |
|
|
Parapoynx (=Nymphula) fluctuosalis Zeller |
Rice caseworm |
|
|
Rupela albinella Cramer |
White stem borer |
|
|
Scirpophaga (=Tryporyza=Schoenobius) incertulas Walker |
Yellow stem borer |
|
|
Scirpophaga innotata Walker |
White stem borer |
|
|
Erebidae |
Rivula atimeta Swinhoe |
Green hairy caterpillar |
|
Noctuidae |
Naranga aenescens Moore |
Green semilooper |
|
Mythimna (=Pseudaletia=Leucania=Cirphis) separata Walker |
Rice ear-cutting caterpillar |
|
|
Mythimna unipuncta Haworth |
True armyworm |
|
|
Sesamia calamistis Hampson |
African pink borer |
|
|
Sesamia inferens Walker |
Pink stem borer |
|
|
Spodoptera frugiperda J.E. Smith |
Fall armyworm |
|
|
Spodoptera litura Fabricius |
Common cutworm |
|
|
Spodoptera mauritia acronyctoides Guenée |
Rice swarming caterpillar |
|
|
Nymphalidae |
Melanitis leda ismene Cramer |
Greenhorned caterpillar |
|
Pyralidae |
Elasmopalpus lignosellus Zeller |
Lesser cornstalk borer |
|
Maliarpha separatella Ragonot |
African white rice borer |
|
|
Coleoptera (beetles) |
||
|
Brachyceridae |
Lissorhoptrus oryzophilus Kuschel |
Rice water weevil |
|
Chrysomelidae |
Dicladispa (=Hispa) armigera Oliver |
Rice hispa |
|
Dicladispa viridicyanea Kraatz |
Rice hispa |
|
|
Oulema (=Lema) oryzae Kuwayama |
Rice leaf beetle |
|
|
Trichispa sericea Guérin-Méneville |
Rice hispa |
|
|
Dryophthoridae |
Echinocnemus squamous Billberg |
Rice root weevil |
|
Scarabaeidae |
Lachnosterna serrata (Fabricius) |
White grub |
|
Diptera (flies) |
||
|
Cecidomyiidae |
Orseolia (=Pachydiplosis) oryzae Wood-Mason |
Rice gall midge |
|
Orseolia oryzivora Harris & Gagné |
Rice gall midge |
|
|
Chloropidae |
Chlorops oryzae Matsumura |
Rice stem maggot |
|
Diopsidae |
Diopsis longicornis Macquart |
Stalk-eyed fly |
|
Diopsis indica Westwood |
Stalk-eyed fly |
|
|
Ephydridae |
Hydrellia griseola Fallén |
Rice leaf miner |
|
Hydrellia philippina Ferino |
Rice whorl maggot |
|
|
Hydrellia sasakii Yuasa & Isitani |
Paddy stem maggot |
|
|
Muscidae |
Atherigona exigua Stein |
Rice seedling maggot |
|
Atherigona oryzae Malloch |
Rice seedling maggot |
|
|
Orthoptera (grasshoppers, locusts and crickets) |
||
|
Acrididae |
Locusta migratoria manilensis Meyen |
Oriental migratory locust |
|
Oxya hyla intricata Stål |
Short-horned grasshopper |
|
|
Oxya japonica japonica Thunberg |
Rice grasshopper |
|
|
Gryllidae |
Euscyrtus concinnus de Haan |
Field cricket |
|
Gryllotalpidae |
Scapteriscus borellii Giglio-Tos |
Mole cricket |
Annex Table 4.C.2. Nematoda
|
Scientific name |
Common name (English) |
|---|---|
|
Aphelenchoides besseyi Christie |
Rice white tip nematode |
|
Ditylenchus angustus (Butler) Filipjev |
Rice stem nematode |
|
Heterodera oryzae Luc & Berdon |
Rice cyst nematode |
|
Hirschmanniella oryzae Luc & Goodey |
Rice root nematode |
|
Meloidogyne graminicola Golden & Birchfield |
Rice root knot nematode |
Annex Table 4.C.3. Mollusca
|
Scientific name |
Common name (English) |
|---|---|
|
Pomacea canaliculata (Lamarck) |
Golden apple snail |
|
Pomacea maculata |
Golden apple snail |
Sources (annex): Ane, N.U. and M. Hussain (2015), “Diversity of insect pests in major rice growing areas of the world”, https://www.semanticscholar.org/paper/Diversity-of-insect-pests-in-major-rice-growing-of-Ane-Hussain/04615420a3c817a37ea6f494574772f84a2a3f09; EPPO (2019), EPPO Global Database, https://gd.eppo.int/ (accessed 11 June 2019); Heinrichs, E.A. and A.T. Barrion (2004), “Rice-feeding insects and selected natural enemies in West Africa: Biology, ecology, identification”, International Rice Research Institute, Los Baños, Philippines, and WARDA–The Africa Rice Center, Abidjan, Côte d’Ivoire; IRRI (n.d.-c), Learn About Best Practices in Rice Farming, http://www.knowledgebank.irri.org/ (accessed 30 October 2020); Joshi and Sebastian (2006), Kyndt, T., D. Fernandez and G. Gheysen (2014), “Plant-parasitic nematode infections in rice: Molecular and cellular insights”, https://www.annualreviews.org/doi/10.1146/annurev-phyto-102313-050111; Nicol, J.M. et al. (2011), “Current nematode threats to world agriculture”, https://doi.org/10.1007/978-94-007-0434-3_2; Pathak, M.D. and Z.R. Khan (1994), Insect Pests of Rice, https://www.cabi.org/isc/abstract/19951100418; Shepard, B.M., A.T. Barrion and J.A. Litsinger (1995), Rice-Feeding Insects of Tropical Asia, International Rice Research Institute, Manila, Philippines, p. 228.
Annex 4.D. Weeds in rice fields
Annex Table 4.D.1. Weeds in rice fields (except for weedy rice (Oryza sativa L. [f. spontanea])
|
Class |
Group |
Species |
|---|---|---|
|
Monocots |
Grasses |
Brachiaria (B. plantaginea, B. platyphylla), Cynodon dactylon, Dactyloctenium aegyptium, Digitaria (D. ciliaris (adscendens), D. sanguinalis, D. setigera), Diplachne fusca, Echinochloa (E. colona, E. crus-galli, E. crus-pavonis, E. glabrescens, E. oryzicola (oryzoides, macrocarpa, phyllopogon)), Eleusine indica, Eragrostis parviflora, Imperata cylindrica, Ischaemum rugosum, Leersia (L. hexandra, L. japonica, L. oryzoides, L. sayanuka), Leptochloa (L. chinensis, L. panicea), Panicum repens, Paspalum (P. distichum, P. paspaloides, P. scrobiculatum), Rottboellia cochinchinensis, Setaria glauca |
|
Sedges |
Bolboschoenus maritimus, Cyperus (C. aromaticus, C. compressus, C. difformis, C. esculentus, C. haspan, C. iria, C. polystachyos, C. rotundus, C. serotinus, C. sphacelatus, C. tenuispica), Eleocharis (E. acicularis, E. acuta, E. congesta, E. dulcis, E. geniculata, E. kuroguwai, E. tetraquetra, E. yokoscensis), Fimbristylis (F. dichotoma, F. diphylla, F. ferruginea, F. littoralis (miliacea)), Schoenoplectiella/Schoenoplectus (S. juncoides, S. mucronatus, S. pungens, S. scirpoides), Scirpus (S. erectus, S. grossus, S. maritimus, S. miliaceus, S. nipponicus, S. planiculmis, S. supinus, S. zeylanica) |
|
|
Floating plants |
Eichhornia crassipes, Lemna (L. minor, L. paucicostata), Pistia stratiotes, Spirodela polyrhiza |
|
|
Others |
Alisma (A. canaliculatum, A. lanceolatum, A. plantago-aquatica), Butomus umbellatus, Commelina (C. benghalensis, C. diffusa), Damasonium minus, Heteranthera (H. limosa, H. reniformis), Limnocharis flava, Monochoria vaginalis, Sagittaria (S. graminea, S. longiloba, S. montevidensis, S. platyphylla, S. pygmaea, S. trifolia), Typha orientalis |
|
|
Dicots |
Aeschynomene (A. aspera, A. indica), Ageratum conyzoides, Alternanthera (A. philoxeroides, A. sessilis), Amaranthus (A. spinosus, A. viridis), Ammannia (A. baccifera, A. multiflora), Bacopa rotundifolia, Celosia argentea, Chromolaena odorata, Eclipta prostrata, Elatine (E. gratioloides, E. triandra), Ipomoea aquatica, Lindernia procumbens, Ludwigia (L. adscendens (stipulacea), L. hyssopifolia, L. octovalvis, L. prostrata (epilobioides)), Lythrum hyssopifolia, Mimosa diplotricha, Oldenlandia corymbosa, Persicaria (Polygonum) hydropiper, Portulaca oleracea, Rotala Indica, Rumex crispus, Sesbania exaltata, Sphenoclea zeylanica, Trianthema portulacastrum |
|
|
Parasites |
Striga (S. asiatica, S. hermonthica) |
|
|
Ferns |
Azolla filiculoides, Marsilea (M. drummondii, M. minuta) |
|
|
Algae |
Blue-green algae |
Anabaena spp., Lyngbya spp., Nostoc spp., Phormidium spp. |
|
Green algae |
Chara spp., Hydrodictyon spp., Pithophora spp., Spirogyra spp. |
Source: IRRI (n.d.-a), How to Control Weeds, http://www.knowledgebank.irri.org/step-by-step-production/growth/weed-management (accessed 2 September 2020); IRRI (n.d.-b), Main Weeds of Rice in Asia, http://www.knowledgebank.irri.org/training/fact-sheets/pest-management/weeds/main-weeds-of-rice-in-asia (accessed 2 September 2020); Kraehmer, H. et al. (2016), “Global distribution of rice weeds - A review”, https://doi.org/10.1016/j.cropro.2015.10.027; Moody, K. (1989), Weeds, Reported in Rice in South and Southeast Asia, International Rice Research Institute, Philippines; Rao, A.N., N. Chandrasena and H. Matsumoto (2017), “Rice weed management in the Asian-Pacific region: An overview”, http://oar.icrisat.org/10210/.
Annex 4.E. Transgenic and genome-edited rice (Oryza sativa)
Transgenic rice
At the dawn of transformation technology research, the production of transgenic rice was more difficult than for other plants. The reason for this was that Agrobacterium does not infect monocotyledons and is not a rice pathogen. Therefore, the transformation of rice required physical methods such as electroporation (Zhang et al., 1988; Shimamoto et al., 1989) and polyethylene glycol method (PEG) (Toriyama et al., 1988; Zhang and Wu, 1988), which are applied to protoplasts. Although transformations in protoplasts have been used, it is quite difficult to regenerate rice plants from them and the occurrence of many somaclonal variations is another serious problem.
The novel transformation method, particle bombardment, was then applied (Christou, Ford and Kofron, 1991). In this method, foreign genes are directly introduced into a callus derived from a cell with high re‑differentiation ability, such as the scutellum. Using this method, the regeneration efficiency was higher than with the protoplasts and the somaclonal variations in the regenerated plants tended to be suppressed when compared to those from the protoplasts.
The next important step was the application of the Agrobacterium method with the Super Binary Vector for the transformation of rice (Hiei et al., 1994). After this research was published, Agrobacterium strains, culture conditions and selection markers were investigated and the transformation efficiency was improved (Toki et al., 2006). Currently, rice is widely used in experiments for monocotyledonous plants as the most easily transformed monocotyledonous crop. The advantages of the Agrobacterium method are its high transformation efficiency and accurate insertion of gene constructs on plasmids when compared to the physical methods, and the number of introduced copies tends to be lower compared to the other methods.
After inserting foreign genes into rice cells/tissues, a selection system for transformed cells was required. An antibiotic resistance gene is generally used as a selection marker. Initially, selection with kanamycin was used with the neomycin phosphotransferase II (NPT-II) gene but the selection efficiency was insufficient and, subsequently, geneticin (G418), to which resistance can be given by the NPT-II gene, was used. As it is a more reliable system, selection based on hygromycin resistance using the hygromycin phosphotransferase (HPT) gene has become more common. Subsequently, selection using glyphosate resistance with the modified 3‑phosphoshikimate-1-carboxyvinyltransferase (mEPSPS) gene, using glufosinate resistance with the phosphinothricin acetyltransferase (Christou, Ford and Kofron, 1991), or using bispyribac salt with the modified acetolactate synthase (mALS) gene have also been developed (Li, Hayashimoto and Murai, 1992). Selection by herbicide tolerance was efficient and produced novel rice not affected by herbicides.
The target of transformation technologies was the insertion of marker genes in order to develop efficient transformation systems in the early research periods but gradually shifted to research for the introduction of practical traits that were being introduced in other crops, such as insect resistance and herbicide resistance. Subsequent research in rice has expanded into other useful agricultural traits such as disease resistance, environmental stress tolerance, high yield and quality improvement. Additionally, functional foods with high contents of useful components or genetically engineered plants for medical use have also been reported. Examples of research reported in the scientific literature are presented in the following paragraphs. No commercial cultivation of these transgenic examples has been conducted at the time of writing (2018).
Disease and pest resistance: In agricultural production, pest resistance traits are of crucial importance. Consequently, significant research effort has been made towards the development of transgenic rice lines exhibiting resistance to agronomically important viruses, rice blast, leaf blight, amongst others (Kathuria et al., 2007). Research on disease resistance mechanisms in rice has led to the identification of multiple pathways involving salicylic acid (SA) (Takatsuji, 2014). Rice transgenic plants overexpressing WRKY45, which is activated by SA signalling, were extremely resistant to rice blast disease, indicating that WRKY45 plays a central role in inducing resistance to filamentous fungi and bacteria (Shimono et al., 2007). In addition, Cry genes (coding for Cry1A, Cry1B, Cry1C, Cry1Ab, and Cry9B) have been introduced to provide resistance to Lepidopteran pests and Galanthus nivalis agglutinin (GNA), which provide resistance to Hemipteran pests including planthoppers (Shabir et al., 2015).
Nutritional change: Rice is one of the most consumed food crops, so supplementing high-nutrient rice could contribute to the improved nutritional status of people in low-nutrition areas. Provitamin A enriched rice, i.e. ‘Golden Rice’, is a notable transgenic example (Ye et al., 2000; Paine et al., 2005). In transgenic research, a soybean ferritin synthesis gene (Vasconcelos et al., 2003) and a human lactoferrin synthesis gene (Nandi et al., 2002) were introduced into rice and shown to enrich its iron and zinc contents respectively. Transgenic rice with enhanced specific amino acid contents such as glycine (Lapitan et al., 2009), lysine (Wu, Chen and Folk, 2003), tryptophan (Tozawa et al., 2001) and cysteine (Lee et al., 2003), have also been reported.
Health functional food or medical use: The development of functional rice to promote human health or transformed rice for medical uses via the induction of intestinal tolerance has been investigated (Wakasa and Takaiwa, 2013; Takaiwa et al., 2015; Shabir et al., 2015). Cedar pollen rice (Takagi et al., 2005), anti‑tick rice (Suzuki et al., 2011) and anti-rheumatic rice (Iizuka et al., 2014) may induce oral immune tolerance and Japanese cedar pollen rice is undergoing clinical research in several hospitals. Examples of the rice lines that may impart health functionalities with functional peptides include blood pressure-regulating rice (Yang et al., 2006; Wakasa et al., 2011) and blood sugar-regulating rice (Sugita et al., 2005). Furthermore, the safety and stability of a rice-based oral vaccine called MucoRice-CTB have also been demonstrated (Azegami et al., 2015).
Efficient transformation capability, in combination with its small diploid genome and the availability of genetic and genomic resources, has led to the use of rice as a model monocotyledonous crop species. Comparative genomics, particularly within the Poaceae family, has contributed to the understanding of cereal crop biology and genome evolution, and advanced crop improvement research and strategies.
Genome-edited rice
Genome editing technologies represent a new era of crop improvement beyond the limits of traditional breeding methods. Among its uses, genome editing can modify specific deoxyribonucleic acid (DNA) sequences at desired positions in the genome, including three categories (Site-Directed Nuclease (SDN)-1, SDN-2 and SDN-3) (Podevin et al., 2013), as well as be applied to induce chromosomal rearrangement (Beying et al., 2020; Schwartz et al., 2020), epigenetic changes and other outcomes that similarly occur in nature. Rice provides an excellent model system for investigating a broad range of agronomically important traits. At the same time, due to the importance of rice as a crop, genome editing technology is expected to be used for modifications that produce commercial traits. A non-exhaustive overview of the results of rice genome editing was compiled mainly with information from scientific papers until 2019 (Mishra, Joshi and Zhao, 2018; Fiaz et al. 2019) (Annex Table 4.E.1).
Yield: Rice yields are increased by improving yield components. Zhang et al. (2017) have previously produced high-yielding rice using knockouts of negative regulators of yield components such as GS3, GS5, DEP1, GW2, Gn1, and TGW6. A co-mutant of GW2, GW5, and TGW6 reported an increase in its 1 000 grain weight of 29.3% (Xu et al., 2016). Heading date is also an important characteristic for yield and heading date has changed significantly in the target mutants of the three major genes (Hd2, Hd4, and Hd5) (Li et al., 2017). Lacchini et al. (2020) attempted to produce dwarf type by modifying HTD1 and increasing the yield by introducing three genes (GN1A, GS3 and GW2).
Disease resistance: The bacterial binding protein in the promoter region of OsSWEET14 was destroyed to confer resistance to leaf blight (Li et al., 2012). Similarly, OsSWEET13 was modified by CRISPR/Cas9 to prevent Transcription Activator-Like (TAL) effects (Zhou et al., 2015). Mutagenesis targeting the ERF transcription factor OsERF922 introduced blast resistance (Wang et al., 2016). Attempts have been made to reduce the infectivity of rice tungro spherical virus (RTSV) and rice tungro bacilliform virus (RTBV) by disrupting the sequences essential for their infection (Macovei et al., 2018). This has implied that plant pest-resistant varieties could be bred by altering essential sequences for plant pest infection or the negative regulators for disease resistance.
Quality and functionality: SBEI and SBEIIb have been removed to produce high-amylose rice (Sun et al., 2017) and high oleic acid rice (Abe et al., 2018) has been produced by disrupting the FAD2 gene.
Herbicide tolerance: Mutations were introduced into the ALS gene to cultivate herbicide-tolerant rice (Li, 2016d; Sun, Y., 2016b). Li et al. (2016d) achieved high tolerance by introducing double point mutations in OsALS. In addition, Sun et al. (2016b) introduced multiple point mutations in the ALS gene by homologous recombination.
Male sterility and fertility restoration: Male-sterile and restorer lines are key materials for hybrid breeding. CRISPR/Cas9 system was applied to develop a rice male-sterile line. A thermo-sensitive genic male sterility was induced by knockout of the TMS5 gene (Zhou et al., 2016; Barman et al., 2019; Li et al., 2019). A photosensitive genic male sterility was also developed by mutagenesis of the CSA gene (Li et al., 2016c). Fertility was restored by a mutation that occurred in the promoter region of RMS gene in the cytoplasmic male-sterile line (Sukemoto, Kazama and Toriyama, 2020). Kazama et al. (2019) demonstrated that TALEN-mediated mutagenesis of the mitochondrial gene, orf79, restored the fertility of cytoplasmic male sterility.
Targeting: Through targeting, via positive-negative selection in addition to gene knockouts, the visualisation of endogenous gene expression will be possible via the knock-in of visible marker genes (Yamauchi et al., 2009). In addition, the precise modification of target genes could be applied to detailed functional analysis and molecular breeding in rice.
Expanding the genome editing toolbox: SpCas9 is widely used but the target site is restricted by the protospacer-adjacent motif (PAM) sequence called -NGG. Therefore, the utilisation of CRISPR/Cpf1 and CaCas9 which have different PAM sequences have been explored. Additionally, SpCas9 was engineered to recognise a single character (-NG) and developed to remove the restriction of PAM sequences consisting of multiple bases (Nishimasu et al., 2018). Furthermore, base editors have been developed as advanced approaches that allow for the direct and irreversible conversion of one target base to another without the need for a double-strand break or donor template (Komor et al., 2016; Nishida et al., 2016). The base editors have already been used in many crops, including rice (Gao, 2021). Genome editing has been successful in rice using a combination of single character PAM recognitions and a base editor (Endo et al., 2019).
Annex Table 4.E.1. List of gene modifications by genome editing in rice (till 2020)
|
Targeted gene |
Strategy |
Molecular functions |
References1 |
|
|---|---|---|---|---|
|
Yield and quality improvement |
LOX3 |
TALENs |
Enhanced storage tolerance |
Ma et al. (2015) |
|
GW2, GW5, and TGW6 |
CRISPR/Cas9 |
Improvement of grain weight |
Xu et al. (2016) |
|
|
Hd2, Hd4, and Hd5 |
CRISPR/Cas9 |
Early maturity of rice varieties |
Li et al. (2017) |
|
|
Gn1a, DEP1, GS3 and IPA1 |
CRISPR/Cas9 |
Improvement of grain number, panicle architecture, grain size, and plant architecture |
Li et al. (2016b) |
|
|
CCD7 |
CRISPR/Cas9 |
Increased tiller number |
Butt et al. (2018) |
|
|
PYLs |
CRISPR/Cas9 |
Improved growth and productivity |
Miao et al. (2018) |
|
|
OsBADH2 |
TALENs |
Enhanced fragrance |
Shan et al. (2015) |
|
|
BADH2 |
CRISPR/Cas9 |
Enhanced fragrance |
Shao et al. (2017) |
|
|
Quality Improvement |
SBE1 and SBEIIb |
CRISPR/Cas9 |
high-amylose rice |
Sun et al. (2017) |
|
OsCYP97A4, OsDSM2, OsCCD4a, OsCCD4b, and OsCCD7 |
CRISPR/Cas9 |
increases -carotene accumulation in rice endosperm |
Yang et al. (2017) |
|
|
OsNramp5 |
CRISPR/Cas9 |
Low Cd-accumulating |
Tang et al. (2017) |
|
|
Biotic stress tolerance |
OsSWEET13 |
TALENs |
Enhanced resistance to bacterial blight |
Li et al. (2012) |
|
OsSWEET13 |
TALENs |
Enhanced resistance to bacterial blight |
Zhou et al. (2015) |
|
|
OsSWEET13 |
TALENs |
Enhanced resistance to bacterial blight |
Blanvillain-Bauf. et al. (2017) |
|
|
Os09g29100 |
TALENs |
Enhanced resistance to bacterial leaf streak |
Cai et al. (2017) |
|
|
OsERF922 |
CRISPR/Cas9 |
Enhanced resistance to blast disease |
Wang et al. (2016) |
|
|
Abiotic stress tolerance |
BEL |
CRISPR/Cas9 |
Herbicide-resistant |
Xu et al. (2014) |
|
OsEPSPS |
CRISPR/Cas9 |
Glyphosate resistant |
Li et al. (2016a) |
|
|
OsALS |
TALENs |
Herbicide-resistant |
Li et al. (2016d) |
|
|
ALS |
CRISPR/Cas9 |
Herbicide-resistant |
Sun et al. (2016) |
|
|
C287 |
Base editing |
Herbicide-resistant |
Shimatani et al. (2017) |
|
|
OsSAPK2 |
CRISPR/Cas9 |
Drought tolerance |
Lou et al. (2017) |
|
|
Nutritional improvement |
OsNRAMP5 |
CRISPR/Cas9 |
Low cadmium content |
Tang et al. (2017) |
|
SBEIIb and SBEI |
CRISPR/Cas9 |
Generation of high-amylose rice |
Sun et al. (2017) |
|
|
OsPDS and OsSBEIIb |
CRISPR/Cpf1 |
carotenoid/starch biosynthesis |
Li et al. (2018) |
|
|
OsFAD2 |
CRISPR/Cas9 |
High oleic/low linoleic |
Abe et al. (2018) |
|
|
GR1, GR2 |
CRISPR/Cas9 |
Carotenoid-enriched |
Dong et al. (2020) |
|
|
Stomatal density |
OsEPFL9 |
CRISPR/Cas9 CRISPR/Cpf1 |
Regulates leaf stomatal density |
Yin et al. (2017) |
|
Nitrogen use efficiency |
NRT1.1Bgene |
Base editing |
Enhance nitrogen use efficiency |
Lu and Zhu (2017) |
|
Senescence and death |
OsCDC48 |
Base editing |
Regulate senescence and death |
Zong et al. (2017) |
|
Hybrid production |
TMS5 |
CRISPR/Cas9 |
Thermo-sensitive male sterility |
Zhou et al. (2016) ; Barman et al. (2019) ; Li et al. (2019) |
|
CSA |
CRISPR/Cas9 |
Photoperiod controlled male-sterile lines |
Li et al. (2016c) |
|
|
RMS |
CRISPR/Cas9 |
Restoration of cytoplasmic male sterility |
Sukemoto, Kazama and Toriyama (2020) |
|
|
Orf79 (mitochondria) |
TALENs |
Restoration of cytoplasmic male sterility |
Kazama et al. (2019) |
Note: This table provides examples of genes that have been modified by genome editing in rice and does not constitute an exhaustive list.
1. Short references are listed in full in the reference section below.
References
Abe, K. et al. (2018), “Production of high oleic/low linoleic rice by genome editing”, Plant Physiology and Biochemistry, Vol. 131, pp. 58-62, https://doi.org/10.1016/j.plaphy.2018.04.033.
Aghaee, M.-A. and L.D. Godfrey (2014), “A century of rice water weevil (Coleoptera: Curculionidae): A history of research and management with an emphasis on the United States”, Journal of Integrated Pest Management, Vol. 5, pp. 1-14, https://doi.org/10.1603/IPM14011.
Akagi, H. et al. (2004), “Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein”, Theoretical and Applied Genetics, Vol. 108, p. 1449-1457, https://doi.org/10.1007/s00122-004-1591-2.
Akagi, H. et al. (1994), “A unique sequence located downstream from the rice mitochondrial atp6 may cause male sterility”, Current Genetics, Vol. 25, p. 52-58, https://doi.org/10.1007/BF00712968.
Akanksha (2009), “Important weeds of rice”, agropedia, http://agropedia.iitk.ac.in/content/important-weeds-rice (accessed 2 September 2020).
Akasaka, M. et al. (2009), “Genetic relationships and diversity of weedy rice (Oryza sativa L.) and cultivated rice varieties in Okayama Prefecture, Japan”, Breeding Science, Vol. 59, p. 401-409, https://doi.org/10.1270/jsbbs.59.401.
American Phytopathological Society (2017), Diseases of Rice (Oryza and Zizania spp.), Common Names of Plant Diseases, https://www.apsnet.org/edcenter/resources/commonnames/Pages/Rice.aspx (accessed 11 June 2019).
Ammiraju, J.S.S. et al. (2006), “The Oryza bacterial artificial chromosome library resources: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza”, Genome Research, Vol.16, pp.140-147, https://dx.doi.org/10.1101%2Fgr.3766306.
Ane, N.U. and M. Hussain (2015), “Diversity of insect pests in major rice growing areas of the world”, Journal of Entomology and Zoology Studies, Vol. 4, pp. 36-41, https://www.semanticscholar.org/paper/Diversity-of-insect-pests-in-major-rice-growing-of-Ane-Hussain/04615420a3c817a37ea6f494574772f84a2a3f09.
Ashikari, M. et al. (2005), “Cytokinin oxidase regulates rice grain production”, Science, Vol. 309, p. 741‑745, https://doi.org/10.1126/science.1113373.
Azegami, K. et al. (1987), “Pseudomonas plantarii sp. nov., the causal agent of rice seedling blight”, International Journal of Systematic Bacteriology, Vol. 37, p. 144-152, https://doi.org/10.1099/00207713-37-2-144.
Azegami, T. et al. (2015), “Novel transgenic rice-based vaccines”, Archivum Immunologiae et Therapiae Experimentalis, Vol. 63, pp. 87-99, https://doi.org/10.1007/s00005-014-0303-0.
Bakti, C. and J. Tanaka (2019), “Detection of dominant QTLs for stigma exsertion ratio in rice derived from Oryza rufipogon accession ‘W0120’”, Breeding Science, Vol. 69, p. 143-150, https://dx.doi.org/10.1270%2Fjsbbs.18139.
Banik, A., S.K. Mukhopadhaya and T.K. Dangar (2016), “Characterization of N2-fixing plant growth promoting endophytic and epiphytic bacterial community of Indian cultivated and wild rice (Oryza spp.) genotypes”, Planta, Vol. 243, pp. 799-812, https://doi.org/10.1007/s00425-015-2444-8.
Barman, H.N. et al. (2019), “Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system”, BMC Plant Biology, Vol. 19, Article 109, https://doi.org/10.1186/s12870-019-1715-0.
Bentolila, S. and S. Stefanov (2012), “A reevaluation of rice mitochondrial evolution based on the complete sequence of male-fertile and male-sterile mitochondrial genomes”, Plant Physiology, Vol. 158, p. 996-1017, https://doi.org/10.1104/pp.111.190231.
Bessho-Uehara, K. et al. (2016), “Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 113, pp. 8969-8974, https://doi.org/10.1073/pnas.1604849113.
Bewley, J.D. et al. (2013), “Germinability during development”, in Seeds: Physiology of Development, Germination and Dormancy, 3rd edition, Springer, pp. 52-59.
Beying, N. et al. (2020), “CRISPR–Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis”, Nature Plants, Vol. 6, pp. 638-645, https://doi.org/10.1038/s41477-020-0663-x.
Bhatia, D. et al. (2017), “Introgression of yield component traits in rice (Oryza sativa ssp. indica) through interspecific hybridization”, Crop Science, Vol. 57, pp. 1557-1573, https://doi.org/10.2135/cropsci2015.11.0693.
Blanvillain-Baufumé, S. et al. (2017), “Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors”, Plant Biotechnology Journal, Vol. 15, pp. 306-317, https://doi.org/10.1111/pbi.12613.
Bowatte, S. et al. (2006), “Molecular analysis of the ammonia oxidizing bacteria community in the surface soil layer of Japanese paddy field”, Soil Science and Plant Nutrition, Vol. 52, pp. 427-431, https://doi.org/10.1111/j.1747-0765.2006.00058.x.
Brar, D.S. and G.S. Khush (2018), “Wild relatives of rice: A valuable genetic resource for genomics and breeding research”, in T.K. Mondal and R.J. Henry (eds.), The Wild Oryza Genomes, pp.1-25, https://doi.org/10.1007/978-3-319-71997-9_1.
Brar, D.S. and G.S. Khush (1997), “Alien introgression in rice”, Plant Molecular Biology, Vol. 35, pp. 35‑47, https://doi.org/10.1023/A:1005825519998.
Brar, D.S. and K. Singh (2011), “Oryza”, in C. Kole (ed.), Wild Crop Relatives: Genomic and Breeding Resources, Cereals, Spring-Verlag, pp. 321-365.
Brar, D.S., R. Elloran and G.S. Khush (1991), “Interspecific hybrids produced through embryo rescue between cultivated and eight wild species of rice”, Rice Genetics Newsletter, Vol. 8, pp. 91-93.
Brar, D.S. et al. (1996), “Gene transfer and molecular characterization of introgression from wild rice Oryza species into rice”, in G.S. Khush (ed.), Rice Genetics III, Internat. Rice Research Instit., Manila, Philippines, pp. 477-486.
Brozynska, M. et al. (2017), “Sequencing of Australian wild rice genomes reveals ancestral relationships with domesticated rice”, Plant Biotechnology Journal, Vol. 15, p. 765-774, https://doi.org/10.1111/pbi.12674.
Burgos, N.R. et al. (2014), “The impact of herbicide-resistant rice technology on phenotypic diversity and population structure of United States weedy rice”, Plant Physiology, Vol. 166, p. 1208-1220, https://doi.org/10.1104/pp.114.242719.
Butt, H. et al. (2018), “Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis”, BMC Plant Biology, Vol. 18, Article 174, https://doi.org/10.1186/s12870-018-1387-1.
CABI (2020), Invasive Species Compendium, https://www.cabi.org/isc/ (accessed 31 August 2020).
Cai, L. et al. (2017), “A transcription activator-like effector Tal7 of Xanthomonas oryzae pv. oryzicola activates rice gene Os09g29100 to suppress rice immunity”, Scientific Reports, Vol. 7, Article 5089, https://doi.org/10.1038/s41598-017-04800-8.
Cartwright, D.W. et al. (1981), “Isolation and characterization of two phytoalexins from rice as momilactones A and B”, Phytochemistry, Vol. 20, pp. 535-537, https://doi.org/10.1016/S0031-9422(00)84189-8.
Cartwright, R.D. et al. (2018), Compendium of Rice Diseases and Pests, 2nd edition, APS Press.
Catling, D. (1992), Rice in Deep Water, International Rice Research Institute, MacMillan Press, London, https://doi.org/10.1007/978-1-349-12309-4.
Caton, B.P. et al. (2010), A Practical Field Guide to Weeds of Rice in Asia, 2nd edition, Internat. Rice Research Instit.
Chang, T.T. (1991), “Findings from a 28-year seed viability experiment”, International Rice Research Newsletter, Vol.16, pp. 5‑6.
Chauhan, B.S. (2013), “Strategies to manage weedy rice in Asia”, Crop Protection, Vol. 48, pp. 51-56, https://doi.org/10.1016/j.cropro.2013.02.015.
Chauhan, B.S. (2012), “Weedy rice (Oryza sativa) II. Response of weedy rice to seed burial and flooding depth”, Weed Science, Vol. 60, pp. 385-388, https://doi.org/10.1614/WS-D-11-00213.1.
Chen, C. et al. (2014), “A two-locus interaction causes interspecific hybrid weakness in rice”, Nature Communications, Vol. 5, pp. 33-57, https://doi.org/10.1038/ncomms4357.
Chen, E. et al. (2019), “The genomics of Oryza species provides insights into rice domestication and heterosis”, Annual Review of Plant Biology, Vol. 70, pp. 639-665, https://doi.org/10.1146/annurev-arplant-050718-100320.
Chen, J. et al. (2013), “Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution”, Nature Communications, Vol. 4, 1595, https://doi.org/10.1038/ncomms2596.
Chen, J. et al. (2008), “A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 105, pp. 11436-11441, https://doi.org/10.1073/pnas.0804761105.
Chen, L.J. et al. (2004), “Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives”, Annals of Botany, Vol. 93, pp. 67-73, https://doi.org/10.1093/aob/mch006.
Chen, S. et al. (2018), “Organic carbon availability limiting microbial denitrification in the deep vadose zone”, Environmental Microbiology, Vol. 20, pp. 980-992, https://doi.org/10.1111/1462-2920.14027.
Chen, X.-P, et al. (2008), “Ammonia-oxidizing archaea: Important players in paddy rhizosphere soil?”, Environmental Microbiology, Vol. 10, pp. 1978-1987, https://doi.org/10.1111/j.1462-2920.2008.01613.x.
Cheng, S.-H. et al. (2007), “Progress in research and development on hybrid rice: A super-domesticate in China”, Annals of Botany, Vol. 100, pp. 959-966, https://doi.org/10.1093/aob/mcm121.
Cho, W.K. et al. (2013), “Current insights into research on Rice stripe virus”, The Plant Pathology Journal, Vol. 29, pp. 223-233, https://doi.org/10.5423/PPJ.RW.10.2012.0158.
Choi, J.Y. et al. (2019), “The complex geography of domestication of the African rice Oryza glaberrima”, PLoS Genetics, Vol. 15, e1007414, https://doi.org/10.1371/journal.pgen.1007414.
Choi, J.Y. et al. (2017), “The rice paradox: Multiple origins but single domestication in Asian rice”, Molecular Biology and Evolution, Vol. 34, pp. 969-979, https://doi.org/10.1093/molbev/msx049.
Chou, C.-H., F.J. Chang and H.I. Oka (1991), “Allelopathic potentials of a wild rice, Oryza perennis”, Taiwania, Vol. 36, pp. 201-210, (in Chinese with English Abstract), https://doi.org/10.6165/tai.1991.36.201.
Christian, M. et al. (2010), “Targeting DNA double-strand breaks with TAL effector nucleases”, Genetics, Vol. 186, pp. 757-761, https://doi.org/10.1534/genetics.110.120717.
Christou, P., T.L. Ford and M. Kofron (1991), “Production of transgenic rice (Oryza sativa L) plants from agronomically important Indica and Japonica varieties via electric discharge particle acceleration of exogenous DNA into immature zygotic embryos”, Nature Biotechnology, Vol. 9, pp. 957-962, https://doi.org/10.1038/nbt1091-957.
Chung, I.-M., S.-J. Hahn and A. Ahmad (2005), “Confirmation of potential herbicidal agents in hulls of rice, Oryza sativa”, Journal of Chemical Ecology, Vol. 31, pp. 1339-1352, https://doi.org/10.1007/s10886-005-5290-5.
Chung, N.-J. and N.-C. Paek (2003), “Photoblastism and ecophysiology of seed germination in weedy rice”, Agronomy Journal, Vol. 95, pp. 184-190, https://doi.org/10.2134/agronj2003.1840.
Cobb, J.N., P.S. Biswas and J.D. Platten (2019), “Back to the future: Revisiting MAS as a tool for modern plant breeding”, Theoretical and Applied Genetics, Vol. 132, pp. 647-667, https://doi.org/10.1007/s00122-018-3266-4.
Copetti, D. and R.A. Wing (2016), “The dark side of the genome: Revealing the native transposable element/repeat content of eukaryotic genomes”, Molecular Plant, Vol. 9, pp. 1664-1666, https://doi.org/10.1016/j.molp.2016.09.006.
Dale, D. (1994), “Insect pests of the rice plant – Their biology and ecology”, in E.A. Heinrichs (ed.), Biology and Management of Rice Insects, Wiley Eastern Ltd, pp. 363-485.
Desaki, Y. et al. (2018), “Plant immunity and symbiosis signaling mediated by LysM receptors”, Innate Immunity, Vol. 24, pp. 92-100, https://doi.org/10.1177%2F1753425917738885.
Dilday, R.H., J. Lin and W. Yan (1994), “Identification of allelopathy in the USDA-ARS germplasm collection”, Australian Journal of Experimental Agriculture, Vol. 34, pp. 907-910, https://doi.org/10.1071/EA9940907.
Dilday, R.H., J.D. Mattice and K.A. Moldenhauer (2000), “An overview of rice allelopathy in the USA”, in Proceedings of International Workshop in Rice Allelopathy, 17-19 August 2000, Kyungpook National University, Taegu, Institute of Agricultural Science and Technology, Kyungpook National University, Korea, pp. 15-26.
Dilday, R.H., P. Nastasi and R.J. Smith Jr. (1989), “Allelopathic observations in rice (Oryza sativa L.) to ducksalad (Heteranthera limosa)”, Proceedings of the Arkansas Academy of Sciences, Vol. 43, pp. 21‑22.
Dilday, R.H. et al. (2001), “Allelopathic potential of rice germplasm against ducksalad, redstem and barnyard grass”, Journal of Crop Production, Vol. 4, pp. 287-301, https://doi.org/10.1300/J144v04n02_11.
Dilday, R.H. et al. (1992), “Weed control with crop allelopathy”, Arkansas Farm Research, Vol. 41, pp. 14‑15.
Dilday, R.H. et al. (1991), “Allelopathic activity in rice (Oryza sativa L.) against ducksalad (Heteranthera limosa (Sw))”, in J.D. Hanson et al. (eds.), Sustainable Agriculture for the Great Plains, Symposium Proceedings, 19‑20 January 1989, USDA, Fort Collins, Colorado, ARS, Springfield, Maryland, pp. 193-201.
Doi, K. et al. (2004), “Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1”, Genes and Development, Vol. 18, pp. 926-936, https://doi.org/10.1101%2Fgad.1189604.
Doi, K., K. Taguchi and A. Yoshimura (1999), “RFLP mapping of S20 and S21 for F1 pollen semi-sterility found in backcross progeny of Oryza sativa and O. glaberrima”, Rice Genetics Newsletter, Vol. 16, pp. 65-68.
Doi, K., K. Taguchi and A. Yoshimura (1998), “A new locus affecting high F1 pollen sterility found in backcross progenies of Japonica rice and African rice”, Rice Genetics Newsletter, Vol. 15, pp. 146‑148.
Dong, O.X. et al. (2020), “Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9”, Nature Communications, Vol. 11, Article 1178, https://doi.org/10.1038/s41467-020-14981-y.
Du, B. et al. (2020), “Current understanding of the genomic, genetic, and molecular control of insect resistance in rice”, Molecular Breeding, Vol. 40, Article 24, https://doi.org/10.1007/s11032-020-1103-3.
Du, B. et al. (2009), “Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 106, pp. 22163-22168, https://doi.org/10.1073/pnas.0912139106.
Du, H. et al. (2017), “Sequencing and de novo assembly of a near complete indica rice genome”, Nature Communications, Vol. 8, 15324, https://doi.org/10.1038/ncomms15324.
Ebana, K. et al. (2001), “Analysis of QTL associated with the allelopathic effect of rice using water-soluble extracts”, Breeding Science, Vol. 51, p. 47-51, https://doi.org/10.1270/jsbbs.51.47.
Ebina, D., M. Nakamura and K. Yamamoto (1998), “Effect of storage period on the germination ability of rice seed”, Bulletin of the Yamaguchi Agricultural Experiment Station, Vol. 49, pp.70-74, (in Japanese with English abstract).
Edwards, J. et al. (2015), “Structure, variation, and assembly of the root-associated microbiomes of rice”, Proceedings of National Academy of Science of the United States of America, Vol. 112, pp. E911‑E920, https://doi.org/10.1073/pnas.1414592112.
Eishire, R.J. et al. (2011), “A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species”, PLoS ONE, Vol. 6, e19379, https://doi.org/10.1371/journal.pone.0019379.
Ellis, R.H., T.D. Hong and E.H. Roberts (1992), “The low-moisture-content limit to the negative logarithmic relation between seed longevity and moisture content in three subspecies of rice”, Annals of Botany, Vol. 69, pp. 53-58, https://doi.org/10.1093/oxfordjournals.aob.a088306.
Elmore, J.M., Z.-J.D. Lin and G. Coaker (2011), “Plant NB-LRR signaling: Upstreams and downstreams”, Current Opinion in Plant Biology, Vol. 14, pp. 365-371, https://doi.org/10.1016/j.pbi.2011.03.011.
Endo, M. et al. (2019), “Genome editing in plants by engineered CRISPR–Cas9 recognizing NG PAM”, Nature Plants, Vol. 5, pp. 14-17, https://doi.org/10.1038/s41477-018-0321-8.
Endo, T. et al. (2009), “Estimate of outcrossing rates in a rice plant (Oryza sativa L.) under field conditions using a purple grain rice cultivar, Okunomurasaki”, Breeding Science, Vol. 59, pp. 195-202, https://doi.org/10.1270/jsbbs.59.195.
EPPO (2019), EPPO Global Database, European and Mediterranean Plant Protection Organization, https://gd.eppo.int/ (accessed 11 June 2019).
Erb, M. and P. Reymond (2019), “Molecular interactions between plants and insect herbivores”, Annual Review of Plant Biology, Vol. 70, pp. 527-557, https://doi.org/10.1146/annurev-arplant-050718-095910.
Fan, C. et al. (2009), “A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker”, Theoretical and Applied Genetics, Vol. 118, pp. 465‑472, https://doi.org/10.1007/s00122-008-0913-1.
FAO (2005), Rice is Life. International Year of Rice 2004 and its Implementation, Food and Agricultural Organisation of the United Nations, Rome.
FAORAP/APSA (2014), Hybrid Rice Development in Asia: Assessment of Limitation and Potential, Proceedings of Expert Consultation, 2-3 July 2014 Bangkok Thailand, FAO Regional Office for Asia and the Pacific on the Food and Agriculture Organization of the United Nations and Asia and Pacific Seed Association.
FAOSTAT (2020), Crop Production Data, Food and Agriculture Organization of the United Nations, Statistics Division online database, https://www.fao.org/faostat/ (accessed 11 March 2022).
Fiaz, S. et al. (2019), “Applications of the CRISPR/Cas9 system for rice grain quality improvement: Perspectives and opportunities”, International Journal of Molecular Sciences, Vol. 20, 888, https://doi.org/10.3390/ijms20040888.
Fuentes, R.R. et al. (2019), “Structural variants in 3000 rice genomes”, Genome Research, Vol. 29, pp. 870-880, http://doi.org/10.1101/gr.241240.118.
Fujii, S. and K. Toriyama (2008), “Genome barriers between nuclei and mitochondria exemplified by cytoplasmic male sterility”, Plant and Cell Physiology, Vol. 49, pp. 1484-1494, https://doi.org/10.1093/pcp/pcn102.
Fujii, Y. (2001), “Screening and future exploitation of allelopathic plants as alternative herbicides with special reference to hairy vetch”, Journal of Crop Production, Vol. 4, pp. 257-275, https://doi.org/10.1300/J144v04n02_09.
Fujii, Y. (1992), “The allelopathic effect of some rice varieties”, in Proceedings of the International Symposium on Biological Control Integrated Management of Paddy and Aquatic Weeds in Asia, 23 October 1992, Tsukuba, Japan, National Agricultural Research Center, pp. 1-6.
Fujii, Y. and S. Hiradate (eds.) (2007), Allelopathy: New Concepts and Methodology, Science Publishers, Inc., Enfield, NH, US.
Fujii, Y. and S. Hiradate (2005), “A critical survey of allelochemicals in action - The importance of total activity and the weed suppression equation”, in Proceedings of the 4th World Congress on Allelopathy, pp. 73-76.
Fujii, Y. and T. Shibuya (1991), “Establishment of a new bioassay specific to allelopathy. Survey of allelopathic plant by the Plant Box Method” (in Japanese), Weed Research (Japan), Vol. 36 (Suppl.), pp. 152-153.
Fujii, Y. et al. (2001), “Screening of allelopathic activity and identification of “Awa-Akamai” (traditional red rice) as the most promising cultivar” (in Japanese), Weed Research (Japan), Vol. 46 (Suppl.), pp. 120-121, https://doi.org/10.3719/weed.46.Supplement_120.
Fujikawa, T. et al. (2012), “Surface α-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants”, PLoS Pathogens, 8, e1002882, https://doi.org/10.1371/journal.ppat.1002882.
Fujino, K. et al. (2010), “Multiple introgression events surrounding the Hd1 flowering-time gene in cultivated rice, Oryza sativa”, Molecular Genetics and Genomics, Vol. 284, pp. 137-146, https://doi.org/10.1007/s00438-010-0555-2.
Fujioka, M. and H. Yoshida (2001), “The potential and problems of agricultural ecosystems for birds in Japan”, Global Environmental Research, Vol. 5, pp. 151-161.
Fujioka, M. et al. (2010), “Bird use of rice fields in Korea and Japan”, Waterbirds, Vol. 33 (Suppl.), pp. 8‑29, https://doi.org/10.1675/063.033.s102.
Fujita, D., A. Kohli and F.G. Horgan (2013), “Rice resistance to planthoppers and leafhoppers”, Critical Reviews in Plant Sciences, Vol. 32, pp. 162-191, https://doi.org/10.1080/07352689.2012.735986.
Fukuoka, S., H. Namai and K. Okuno (1998), “Geographical variation of the genes controlling hybrid breakdown and genetic differentiation of the chromosomal regions harboring these genes in Asian cultivated rice, Oryza sativa L.”, Genes and Genetic Systems, Vol. 73, pp. 211-217, https://doi.org/10.1266/ggs.73.211.
Fuller, D.Q. et al. (2009), “The domestication process and domestication rate in rice: Spikelet bases from the Lower Yangtze”, Science, Vol. 323, pp. 1607-1610, http://doi.org/10.1126/science.1166605.
Gao, C. (2021), “Genome engineering for crop improvement and future agriculture”, Cell, Vol. 184, pp. 1621-1635, https://doi.org/10.1016/j.cell.2021.01.005.
Garavito, A. et al. (2010), “A genetic model for the female sterility barrier between Asian and African cultivated rice species”, Genetics, Vol. 185, pp. 1425-1440, https://doi.org/10.1534/genetics.110.116772.
Gasiunas, G. et al. (2012), “Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 109, E2579-E2586, https://doi.org/10.1073/pnas.1208507109.
Ge, S. et al. (1999), “Phylogeny of rice genomes with emphasis on origins of allotetraploid species”, Proceeding of the National Academy of Science of the United States of America, Vol. 96, pp. 14400‑14405, https://doi.org/10.1073/pnas.96.25.14400.
GRiSP (2013), Rice Almanac, 4th edition, Global Rice Science Partnership, International Rice Research Institute, Los Baos, Philippines.
Grist, D.H. and R.J.A.W. Lever (1969), Pests of Rice, Longmans, London, and Green and Co., Ltd. Harlow.
Gu, B. et al. (2015), “An-2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice”, Molecular Plant, Vol. 8, pp. 1635-1650, https://doi.org/10.1016/j.molp.2015.08.001.
Gu, Y., P. Wang and C.H. Kong (2009), “Urease, invertase, dehydrogenase and polyphenoloxidase activities in paddy soil influenced by allelopathic rice variety”, European Journal of Soil Biology, Vol. 45, pp. 436-441, https://doi.org/10.1016/j.ejsobi.2009.06.003.
Gu, Y., P. Wang and C.H. Kong (2008), “Effects of rice allelochemicals on the microbial community of flooded paddy soil”, Allelopathy Journal, Vol. 22, pp. 299-309.
Guo, J. et al. (2018), “Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice”, Nature Genetics, Vol. 50, pp. 297-306, https://doi.org/10.1038/s41588-018-0039-6.
Guo, Y. et al. (2009), “Allelopathic effects of rice cultivars on barnyardgrass growth to reduce the herbicide dose”, Allelopathy Journal, Vol. 24, pp. 321-329.
Gurney, A.L. et al. (2006), “A novel form of resistance in rice to the angiosperm parasite Striga hermonthica”, New Phytologist, Vol. 169, pp. 199-208, https://doi.org/10.1111/j.1469-8137.2005.01560.x.
Gutjahr, C., L. Casieri and U. Paszkowski (2009), “Glomus intraradices induces changes in root system architecture of rice independently of common symbiosis signaling”, New Phytologist, Vol. 182, pp. 829-837, https://doi.org/10.1111/j.1469-8137.2009.02839.x.
Hajiboland, R., N. Aliasgharzad and R. Barzeghar (2009), “Phosphorus mobilization and uptake in mycorrhizal rice (Oryza sativa L.) plants under flooded and non-flooded conditions”, Acta agriculturae Slovenica, Vol. 93, pp. 153-161, https://doi.org/10.2478/V10014-009-0010-4.
Harsonowati, W., R.I. Astuti and A.T. Wahyudi (2017), “Leaf blast disease reduction by rice-phyllosphere actinomyecetes producing bioactive compounds”, Journal of General Plant Pathology, Vol. 83, pp. 98‑108, https://doi.org/10.1007/s10327-017-0700-4.
Hattori, M. (2001), “Probing behavior of the brown planthopper, Nilaparvata lugens Stål (Homoptera: Delphacidae) on a non-host barnyard grass, and resistant and susceptible varieties of rice”, Applied Entomology and Zoology, Vol. 36, pp. 83-89, https://doi.org/10.1303/aez.2001.83.
Hattori, M. (1997), “Feeding behavior of the green rice leafhopper, Nephotettix cincticeps (Homoptera: Cicadellidae) towards pure phloem sap collected from resistant and susceptible rice varieties”, Applied Entomology and Zoology, Vol. 32, pp. 409-412, https://doi.org/10.1303/aez.32.409.
Heinrichs, E.A. (ed.) (1994) Biology and Management of Rice Insects, Wiley Eastern Ltd.
Heinrichs, E.A. and A.T. Barrion (2004), Rice-feeding Insects and Selected Natural Enemies in West Africa: Biology, Ecology, Identification, International Rice Research Institute, Los Baños, Philippines, and WARDA–The Africa Rice Center, Abidjan, Côte d’Ivoire.
Heinrichs, E.A., F.G. Medano and H.R. Rapusas (1985), Genetics Evaluation for Insect Resistance in Rice, International Rice Research Institute, Manila, Philippines.
Hibino, H. et al. (1986a), “Rice ragged stunt virus”, Technical Bulletin of the Tropical Agriculture Research Center, Vol. 21, pp. 14-33.
Hibino, H. et al. (1986b), “Rice grassy stunt virus – Purification and serology –”, Technical Bulletin of the Tropical Agriculture Research Center, Vol. 21, p. 34-40.
Hibino, H. (1983), “Relations of rice tungro bacilliform and rice tungro spherical viruses with their vector Nephotettix virescens”, Annals of Phytopathological Society of Japan, Vol. 49, pp. 545-553, https://doi.org/10.3186/jjphytopath.49.545.
Hiei, Y. et al. (1994), “Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of' the boundaries of the T-DNA”, The Plant Journal, Vol. 6, pp. 271-282, https://doi.org/10.1046/j.1365-313X.1994.6020271.x.
Hiradate, S. (2006), “Isolation strategies for finding bioactive compounds: Specific activity vs. total activity”, Natural Products for Pest Management, ACS Symposium Series, Vol. 927, pp. 113-126, https://doi.org/10.1021/bk-2006-0927.ch009.
Hiradate, S. et al. (2010), “Quantitative evaluation of allelopathic potentials in soils: Total activity approach”, Weed Science, Vol. 58, pp. 258-264, https://doi.org/10.1614/WS-D-09-00085.1.
Hiraguri, A. et al. (2010), “Complete sequence analysis of rice transitory yellowing virus and its comparison to rice yellow stunt virus”, Archives of Virology, Vol. 155, pp. 243-245, https://doi.org/10.1007/s00705-009-0557-8.
Honda, K. et al. (2007), “Retention of Rice dwarf virus by descendants of pairs of viruliferous vector insects after rearing for 6 years”, Phytopathology, Vol. 97, pp. 712-716, https://doi.org/10.1094/PHYTO-97-6-0712.
Hong, W.-J. et al. (2019), “Infrastructures of systems biology that facilitate functional genomic study in rice”, Rice, Vol. 12, Article 15, https://doi.org/10.1186/s12284-019-0276-z.
Hori, K., K. Matsubara and M. Yano (2016), “Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics”, Theoretical and Applied Genetics, Vol. 129, pp. 2241-2252, https://doi.org/10.1007/s00122-016-2773-4.
Hoshikawa, K. (1989), “Chapter X: Heading, anthesis and fertilization”, in The Growing Rice Plant, Nobunkyo, Tokyo, pp. 237-253.
Hosoi, J. et al. (2010), “Viability dynamics of overwintering seeds of weedy rice in Nagano Prefecture placed on soil surface and buried in soil”, Japanese Journal of Crop Science, Vol. 79, pp. 322-326, https://doi.org/10.1626/jcs.79.322.
Hou, J. et al. (2019), “ESA1 is involved in embryo sac abortion in interspecific hybrid progeny of rice”, Plant Physiology, Vol. 180, pp. 356-366, https://doi.org/10.1104/pp.18.01374.
Howard, R.J. and B. Valent (1996), “Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea”, Annual Review of Microbiology, Vol. 50, pp. 491-512, https://doi.org/10.1146/annurev.micro.50.1.491.
Hu, F. et al. (2006), “Molecular mapping of a pollen killer gene S29(t) in Oryza glaberrima and co-linear analysis with S22 in O. glumaepatula”, Euphytica, Vol. 151, pp. 273-278, https://doi.org/10.1007/s10681-006-9146-z.
Hu, F.Y. et al. (2003), “Convergent evolution of perenniality in rice and sorghum”, Proceeding of the National Academy of Sciences of the United States of America, Vol. 100, pp. 4050-4054, https://doi.org/10.1073/pnas.0630531100.
Hu, L. et al. (2017), “The coiled-coil and nucleotide binding domains of BROWN PLANTHOPPER RESISTANCE14 function in signaling and resistance against planthopper in rice”, The Plant Cell, Vol. 29, pp. 3157-3185, https://doi.org/10.1105/tpc.17.00263.
Hua, L. et al. (2015), “LABA1, a domestication gene associated with long, barbed awns in wild rice”, The Plant Cell, Vol. 27, pp. 1875-1888, https://doi.org/10.1105/tpc.15.00260.
Huang, H.J. et al. (2016), “Screening and functional analyses of Nilaparvata lugens salivary proteome”, Journal of Proteome Research, Vol. 15, pp. 1883-1896, https://doi.org/10.1021/acs.jproteome.6b00086.
Huang, J.Z. et al. (2014), “Workable male sterility systems for hybrid rice: Genetics, biochemistry, molecular biology, and utilization”, Rice, Vol. 7, Article 13, https://doi.org/10.1186/s12284-014-0013-6.
Huang, T. et al. (2007), “Genetic analysis and mapping of genes involved in fertility of Pingxiang dominant genic male sterile rice”, Journal of Genetics and Genomics, Vol. 34, pp. 616-622, https://doi.org/10.1016/S1673-8527(07)60070-8.
Huang, W. et al. (2015), “Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility”, Proceedings of the National Academy of Sciences, Vol. 112, pp. 14984-14989, https://doi.org/10.1073/pnas.1511748112.
Huang, X. et al. (2012), “A map of rice genome variation reveals the origin of cultivated rice”, Nature, Vol. 490, pp. 497-501. https://doi.org/10.1038/nature11532.
Huang, X. et al. (2010), “Genome-wide association studies of 14 agronomic traits in rice landraces”, Nature Genetics, Vol. 42, pp. 961-967, https://doi.org/10.1038/ng.695.
Huang, X. et al. (2009), “High-throughput genotyping by whole-genome resequencing”, Genome Research, Vol. 19, pp. 1068-1076, http://www.genome.org/cgi/doi/10.1101/gr.089516.108.
Ichitani, K. et al. (2011), “Chromosomal location of HWA1 and HWA2, complementary hybrid weakness genes in rice”, Rice, Vol. 4, pp. 29-38, https://doi.org/10.1007/s12284-011-9062-2.
ICTV (2019), International Committee on Taxonomy of Viruses, https://talk.ictvonline.org (accessed 11 June 2019).
Iizuka, M. et al. (2014), “Suppression of collagen-induced arthritis by oral administration of transgenic rice seeds expressing altered peptide ligands of type II collagen”, Plant Biotechnology Journal, Vol. 12, pp. 1143-1152, https://doi.org/10.1111/pbi.12223.
Ikeda, K. et al. (2007), “Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate”, The Plant Journal, Vol. 51, pp. 1030-1040, https://doi.org/10.1111/j.1365-313X.2007.03200.x.
Ikehashi, H. and H. Araki (1986), “Genetics of F1 sterility in remote crosses of rice”, Rice Genetics, Vol. 1, pp. 119-130, https://doi.org/10.1142/9789812814265_0011.
Imchen, M. et al. (2019), “16S rRNA gene amplicon based metagenomic signatures of rhizobiome community in rice field during various growth stages”, Frontiers in Microbiology, Vol. 10, 2103, https://doi.org/10.3389/fmicb.2019.02103.
Index Fungorum (2019), The Global Database of Fungal Names, http://www.indexfungorum.org/ (accessed 11 June 2019).
International Rice Genome Sequencing Project (2005), “The map-based sequence of the rice genome”, Nature, Vol. 436, pp. 793-800, https://doi.org/10.1038/nature03895.
IRRI (2016), Rice Diseases: Biology and Selected Management Practices, International Rice Research Institute, http://rice-diseases.irri.org/home/contents.
IRRI (2012), The International Rice Genebank – Conserving Rice, International Rice Research Institute, https://web.archive.org/web/20121023054703/http:/irri.org/index.php?option=com_k2&view=item&id=9960&lang=en.
IRRI (1985), International Rice Research: 25 Years of Partnership, IRRI press, Manila, pp. 1-188.
IRRI (1983), Field Problems of Tropical Rice, Revised edition, International Rice Research Institute.
IRRI (n.d.-a), How to Control Weeds, Rice Knowledge Bank, International Rice Research Institute, http://www.knowledgebank.irri.org/step-by-step-production/growth/weed-management (accessed 2 September 2020).
IRRI (n.d.-b), Main Weeds of Rice in Asia, Rice Knowledge Bank, International Rice Research Institute, http://www.knowledgebank.irri.org/training/fact-sheets/pest-management/weeds/main-weeds-of-rice-in-asia (accessed 2 September 2020).
IRRI (n.d.-c), Learn About Best Practices in Rice Farming, Rice Knowledge Bank, International Rice Research Institute, http://www.knowledgebank.irri.org/ (accessed 30 October 2020).
Ishii, T., et al. (1994), “Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice, O. sativa”, Genome, Vol. 37, pp. 217-221, https://doi.org/10.1139/g94-030.
Ishikawa, R. et al. (2006), “Genetic erosion from modern varieties into traditional upland rice cultivars (Oryza sativa L.) in northern Thailand”, Genetic Resources and Crop Evolution, Vol. 53, pp. 245-252, https://doi.org/10.1007/s10722-004-6132-y.
Ishikawa, R. et al. (2002a), “Origin of cytoplasm substituted rice cultivars found in Japan”, Theoretical and Applied Genetics, Vol. 105, pp. 608-613, https://doi.org/10.1007/s00122-002-0898-0.
Ishikawa, R. et al. (2002b), “Different maternal origins of Japanese lowland and upland rice populations”, Theoretical and Applied Genetics, Vol. 104, pp. 976-980, https://doi.org/10.1007/s00122-001-0807-y.
ISSG (n.d.), View 100 of the World’s Worst Invasive Alien Species, Invasive Species Specialist Group, http://www.issg.org/worst100_species.html (accessed 31 August 2020).
Isshiki, M. et al. (1998), “A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5’ splice site of the first intron”, The Plant Journal, Vol. 15, pp.133-138, https://doi.org/10.1046/j.1365-313X.1998.00189.x.
Itabashi, E., T. Kazama and K. Toriyama (2009), “Characterization of cytoplasmic male sterility of rice with lead rice cytoplasm in comparison with that with Chinsurah Boro II cytoplasm”, Plant Cell Reports, Vol. 28, pp. 233-239, https://doi.org/10.1007/s00299-008-0625-7.
Itabashi, E. et al. (2011), “The fertility restorer gene, Rf2, for lead rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein”, The Plant Journal, Vol. 65, pp. 359-367, https://doi.org/10.1111/j.1365-313X.2010.04427.x.
Itoh, H. et al. (2018), “Genomic adaptation of flowering-time genes during the expansion of rice cultivation area”, The Plant Journal, Vol. 94, pp. 895-909, https://doi.org/10.1111/tpj.13906.
Iwabuchi, M., J. Kyozuka and K. Shimamoto (1993), “Processing followed by complete editing of an altered mitochondrial atp6 RNA restores fertility of cytoplasmic male sterile rice”, The EMBO Journal, Vol. 12, pp. 1437-1446, https://doi.org/10.1002/j.1460-2075.1993.tb05787.x.
Jabran, K. (2017), “Rice allelopathy for weed control”, in K. Jabran. (ed.), Manipulation of Allelopathic Crops for Weed Control, Springer International Publishing AG, Switzerland, pp. 35-47, https://doi.org/10.1007/978-3-319-53186-1_5.
Jacquemin, J. et al. (2013), “The International Oryza Map Alignment Project: Development of a genus-wide comparative genomics platform to help solve the 9 billion-people question”, Current Opinion in Plant Biology, Vol. 16, pp. 147-156, https://doi.org/10.1016/j.pbi.2013.02.014.
Japanese Society of Applied Entomology and Zoology (2006), Major Insect and Other Pests of Economic Plants in Japan, Revised edition, Tokyo.
Jena, K.K. (2010), “The species of the genus Oryza and transfer of useful genes from wild species into cultivated rice, O. sativa”, Breeding Science, Vol. 60, pp. 518-523, https://doi.org/10.1270/jsbbs.60.518.
Jena, K.K. (1994) “Production of intergeneric hybrid between Oryza sativa L. and Porteresia coarctata T”, Current Science, Vol. 67, pp. 744-746, http://www.jstor.org/stable/24095851.
Jena, K.K. and G.S. Khush (1990), “Introgression of genes from Oryza offficinalis Well ex Watt into cultivated rice, O. sativa L.”, Theoretical and Applied Genetics, Vol. 80, pp. 737-745, https://doi.org/10.1007/BF00224186.
Jena, K.K. and D.J. Mackill (2008), “Molecular markers and their use in marker-assisted selection in rice”, Crop Science, Vol. 48, pp. 1266-1276, https://doi.org/10.2135/cropsci2008.02.0082.
Jiao, Y. et al. (2010), “Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice”, Nature Genetics, Vol. 42, pp. 541-544, https://doi.org/10.1038/ng.591.
Jin, J. et al. (2008), “Genetic control of rice plant architecture under domestication”, Nature Genetics, Vol. 40, pp.1365-1369, https://doi.org/10.1038/ng.247.
Jin, X. et al. (2017), “Introgression from cultivated rice alters genetic structures of wild relative populations: Implications for in situ conservation”, AOB Plants, Vol. 10, plx055, https://doi.org/10.1093/aobpla/plx055.
Jinek, M. et al. (2012), “A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity”, Science, Vol. 337, pp. 816-821, http://science.sciencemag.org/content/337/6096/816.
Jonson, G.B. et al. (2020), “Reemerging rice orange leaf phytoplasma with varying symptoms expressions and its transmission by a new leafhopper vector—Nephotettix virescens Distant”, Pathogens, Vol. 9, p. 990, https://doi.org/10.3390/pathogens9120990.
Joseph, L., P. Kuriachan and G. Thomas, (2008), “Is Oryza malampuzhaensis Krish. et Chand. (Poaceae) a valid species? Evidence from morphological and molecular analyses”, Plant Systematics and Evolution, Vol. 270, pp. 75-94, https://doi.org/10.1007/s00606-007-0606-2.
Joshi, R.C. and L.S. Sebastian (2006), Global Advances in Ecology and Management of Golden Apple Snails, Philippine Rice Research Institute, Nueva Ecija, Philippines.
Joshi, R.C., A.T. Barrion and L.S. Sebastian (eds.) (2007), “Rice black bugs: Taxonomy, ecology, biology, and management of invasive species”, Philippines Rice Research Institute, p. 793.
Juliano, B.O. (1985), “Polysaccharides, proteins, and lipid of rice”, in B.O. Juliano (ed.), Rice: Chemistry and Technology, 2nd edition, American Association Cereal Chemists, St. Paul, Minnesota, pp 59-174.
Juliano, B.O. and C.P. Villareal (1993), Grain Quality Evaluation of World Rices, International Rice Research Institute (IRRI), p. 214.
Juliano B.O., C.M. Perez and T.T. Chang (1990), “Varietal differences in longevity of tropical rough rice stored under ambient conditions”, Seed Science and Technology, Vol. 18, pp. 361-369.
Juliano, B.O. et al. (1968), “Screening for high protein rice varieties”, Cereal Science Today, Vol. 13, pp. 299-301.
Kadota, I. and A. Ohuchi (1990), “Symptoms and ecology of bacterial brown stripe of rice”, Japan Agricultural Research Quarterly, Vol. 24, pp. 15-21.
Kalia, S. and R. Rathour (2019), “Current status on mapping of genes for resistance to leaf- and neck-blast disease in rice”, 3 Biotech., Vol. 9, p. 209, https://doi.org/10.1007/s13205-019-1738-0.
Kaloshian, I. and L.L. Walling (2016), “Hemipteran and dipteran pests: Effectors and plant host immune regulators”, Journal of Integrative Plant Biology, Vol. 58, pp. 350-361, https://doi.org/10.1111/jipb.12438.
Kaneda, C. (1993), “Rice”, in Traditional Crop Breeding Practices: An Historical Review to Serve as a Baseline for Assessing the Role of Modem Biotechnology, OECD, Paris, pp. 37-46.
Kathuria, H. et al. (2007), “Advances in transgenic rice biotechnology”, Critical Reviews in Plant Sciences, Vol. 26, pp. 65-103, https://doi.org/10.1080/07352680701252809.
Kato, H. and H. Namai (1987), “Floral characteristics and environmental factors for increasing natural out crossing rate for F1 hybrid seed production of rice Oryza sativa L.”, Japanese Journal of Breeding, Vol. 37, pp. 318–330, https://doi.org/10.1270/jsbbs1951.37.318 (in Japanese with English abstract).
Kato, T. et al. (1973), “Momilactones are growth inhibitors from rice, Oryza sativa L.”, Tetrahedron Letters, Vol. 39, pp. 3861-3864, https://doi.org/10.1016/S0040-4039(01)87058-1.
Kato-Noguchi, H. and R.J. Peters (2013), “The role of momilactones in rice allelopathy”, Journal of chemical Ecology, Vol. 39, pp. 175-185, https://doi.org/10.1007/s10886-013-0236-9.
Kato-Noguchi, H. and T. Ino (2005), “Possible involvement of momilactone B in rice allelopathy”, Journal of Plant Physiology, Vol. 162, pp. 718-21, https://doi.org/10.1016/j.jplph.2004.11.009.
Kato-Noguchi, H. and T. Ino (2003), “Rice seedlings release momilactone B into the environment”, Phytochemistry, Vol. 63, pp. 551-554, https://doi.org/10.1016/S0031-9422(03)00194-8.
Kawabe, S. (1985), “Mechanism of varietal resistance to the rice green leafhopper (Nephotettix cincticeps Uhler)”, Japan Agricultural Research Quarterly, Vol. 19, pp. 115-124.
Kawahara, Y. et al. (2013), “Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data”, Rice, Vol. 6/4, https://doi.org/10.1186/1939-8433-6-4.
Kawasaki, A. et al. (2009), “Molecular constitution of weedy rice (Oryza sativa L.) found in Okayama prefecture, Japan”, Breeding Science, Vol. 59, pp. 229-236, https://doi.org/10.1270/jsbbs.59.229.
Kazama, T. and K. Toriyama (2014), “A fertility restorer gene, Rf4, widely used for hybrid rice breeding encodes a pentatricopeptide repeat protein”, Rice, Vol. 7, Article 28, https://doi.org/10.1186/s12284-014-0028-z.
Kazama, T. and K. Toriyama (2003), “A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice”, FEBS Letters, Vol. 544, pp. 99‑102, https://doi.org/10.1016/S0014-5793(03)00480-0.
Kazama, T. et al. (2019), “Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing”, Nature Plants, Vol. 5, pp. 722-730, https://doi.org/10.1038/s41477-019-0459-z.
Kazama, T. et al. (2016), “Mitochondrial ORF79 levels determine pollen abortion in cytoplasmic male sterile rice”, The Plant Journal, Vol. 85, pp. 707-716, https://doi.org/10.1111/tpj.13135.
Khan, Z.R. et al. (1990), “World Bibliography of Rice Stem Borers 1794-1990”, International Rice Research Institute.
Khanh T.D., T.D. Xuan and I.M. Chung (2007), “Rice allelopathy and the possibility for weed management”, Annals of Applied Biology, Vol. 151, pp. 325-339, https://doi.org/10.1111/j.1744-7348.2007.00183.x.
Khatun, S. and T.J. Flowers (1995), “The estimation of pollen viability in rice”, Journal of Experimental Botany, Vol. 46, pp. 151-154, www.jstor.org/stable/23694857.
Khush, G.S (1997), “Origin, dispersal, cultivation and variation of rice”, Plant Molecular Biology, Vol. 35, pp. 25–34.
Khush, G.S. (1977), “Disease and insect resistance in rice”, Advances in Agronomy, Vol. 29, pp. 265‑341, https://doi.org/10.1016/S0065-2113(08)60221-7.
Khush, G.S. and D.S. Brar (2017), “Alien introgression in rice”, Nucleus, Vol. 60, pp. 251-261, https://doi.org/10.1007/s13237-017-0222-7.
Khush, G.S. and D.S. Brar (1992), “Overcoming barriers in hybridization”, in G. Kalloo and J.B. Chowdhury (eds.), Distant Hybridization of Crop Plants, Monographs on Theoretical and Applied Genetics, Vol. 16, pp. 47-61, https://doi.org/10.1007/978-3-642-84306-8_4.
Khush, G.S., W.R. Coffman and H.M. Beachell (2001), “The history of rice breeding: IRRI’s contribution”, in W.G. Rockwood (ed.), Rice Research and Production in the 21st Century: Symposium Honoring Robert F. Chandler, Jr., International Rice Research Institute, Los Baños, Philippines, pp. 117-135, http://books.irri.org/9712201635_content.pdf.
Khush, G.S. et al. (1994), “Apomixis for rice improvement”, in G.S. Khush (ed.), Apomixis: Exploiting Hybrid Vigor in Rice, pp. 1-21.
Kim, K. et al. (2015), “Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species”, Scientific Reports, Vol. 5, 15655, https://doi.org/10.1038/srep15655.
Kim, Y.-J. and D. Zhang (2018), “Molecular control of male fertility for crop hybrid breeding”, Trends in Plant Science”, Vol. 23, pp. 53-65, https://doi.org/10.1016/j.tplants.2017.10.001.
Kiritani, K. (2007), “Implications of an unintended area-wide IPM for Chilo suppressalis in Japan”, Extension Bulletin 588, Food and Fertilizer Technology Center, pp. 1-9.
Kishimoto, K. et al. (2010), “Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice”, The Plant Journal, Vol. 64, pp. 343-354, https://doi.org/10.1111/j.1365-313X.2010.04328.x.
Kobayashi, K., Y. Yogo and H. Sugiyama (1995), “Differential growth response of rice cultivars to pyrazosulfuron-ethyl”, Journal of Weed Science and Technology, Vol. 40, pp. 104-109, https://doi.org/10.3719/weed.40.104.
Koga, Y. et al. (1971), “Studies on the longevity of pollen grains of rice, Oriza sativa L., I. Morphological change of pollen grains after shedding”, Cytologia, Vol. 36, pp. 104-110, https://doi.org/10.1508/cytologia.36.104.
Kögel-Knabner, I. et al. (2010), “Biogeochemistry of paddy soils”, Geoderma, Vol. 157, pp. 1-14, https://doi.org/10.1016/j.geoderma.2010.03.009.
Köhler, F.E. (1897), Köhler’s Medizinal-Pflanzen.
Komor, A.C. et al. (2016), “Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage”, Nature, Vol. 533, pp. 420-424, https://doi.org/10.1038/nature17946.
Komori, T. et al. (2004), “Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.)”, The Plant Journal, Vol. 37, pp. 315-325, https://doi.org/10.1046/j.1365-313X.2003.01961.x.
Konishi, S., K. Ebana and T. Izawa (2008), “Inference of the japonica rice domestication process from the distribution of six functional nucleotide polymorphisms of domestication-related genes in various landraces and modern cultivars”, Plant and Cell Physiology, Vol. 49, pp. 1283-1293, https://doi.org/10.1093/pcp/pcn118.
Konishi, S. et al. (2006), “An SNP caused loss of seed shattering during rice domestication”, Science, Vol. 312, pp. 1392-1396, https://doi.org/10.1126/science.1126410.
Kovach, M.J., M.T. Sweeney and S.R. McCouch (2007), “New insights into the history of rice domestication”, Trends in Genetics, Vol. 23, pp. 578-587, https://doi.org/10.1016/j.tig.2007.08.012.
Kraehmer, H. et al. (2016), “Global distribution of rice weeds - A review”, Crop Protection, Vol. 80, pp. 73-86, https://doi.org/10.1016/j.cropro.2015.10.027.
Kubo, T. and A. Yoshimura (2005), “Epistasis underlying female sterility detected in hybrid breakdown in a Japonica-Indica cross of rice (Oryza sativa L.)”, Theoretical and Applied Genetics, Vol. 110, pp. 346-355, https://doi.org/10.1007/s00122-004-1846-y.
Kubo, T. and A. Yoshimura (2002), “Genetic basis of hybrid breakdown in a Japonica/Indica cross of rice, Oryza sativa L.”, Theoretical and Applied Genetics, Vol. 105, pp. 906-911, https://doi.org/10.1007/s00122-002-1059-1.
Kubo, T. et al. (2016), “Two tightly linked genes at the hsa1 locus cause both F1 and F2 hybrid sterility in rice”, Molecular Plant, Vol. 9, pp. 221-232, https://doi.org/10.1016/j.molp.2015.09.014.
Kubo, T. et al. (2008), “A novel epistatic interaction at two loci causing hybrid male sterility in an inter-subspecific cross of rice (Oryza sativa L.)”, Genes and Genetic Systems, Vol. 83, pp. 443-453, https://doi.org/10.1266/ggs.83.443.
Kuboyama, T. et al. (2009), “Fine mapping of HWC2, a complementary hybrid weakness gene, and haplotype analysis around the locus in rice”, Rice, Vol. 2, pp. 93-103, https://doi.org/10.1007/s12284-009-9026-y.
Kurata, N. and T. Omura (1984), “Chromosome analysis”, Developments in Crop Science, Vol. 7, pp. 305-320, https://doi.org/10.1016/B978-0-444-99615-2.50017-X.
Kuwatsuka, S. and H. Shindo (1973), “Behavior of phenolic substances in the decaying process of plants. I. Identification and quantitative determination of phenolic acids in rice straw and its decayed product by gas chromatography”, Soil Science and Plant Nutrition, Vol. 19, pp. 219–227, https://doi.org/10.1080/00380768.1973.10432591.
Kyndt, T., D. Fernandez and G. Gheysen (2014), “Plant-parasitic nematode infections in rice: Molecular and cellular insights”, Annual Review of Phytopathology, Vol. 52, pp. 135-153, https://www.annualreviews.org/doi/10.1146/annurev-phyto-102313-050111.
Lacchini, E. et al. (2020), “CRISPR-mediated accelerated domestication of African rice landraces”, PLoS One, Vol. 15, e0229782, https://doi.org/10.1371/journal.pone.0229782.
Lane, S.J., A. Azuma and H. Higuchi (1998), “Wildfowl damage to agriculture in Japan”, Agriculture, Ecosystems and Environment, Vol. 70, pp. 69-77, https://doi.org/10.1016/S0167-8809(98)00114-5.
Lapitan, V.C. et al. (2009), “Mapping of quantitative trait loci using a doubled-haploid population from the cross of Indica and Japonica cultivars of rice”, Crop Science, Vol. 49, pp. 1620-1628, https://doi.org/10.2135/cropsci2008.11.0655.
Lee, H.S. et al. (2010), “Identification of molecular markers for photoblastism in weedy rice”, Korean Journal of Breeding Science, Vol. 42, pp. 144-150.
Lee, T.T.T. et al. (2003), “Enhanced methionine and cysteine levels in transgenic rice seeds by the accumulation of sesame 2S albumin”, Bioscience, Biotechnology, and Biochemistry, Vol. 67, p. 1699‑1705, https://doi.org/10.1271/bbb.67.1699.
Li, C., A. Zhou and T. Sang (2006), “Rice domestication by reducing shattering”, Science, Vol. 311, pp. 1936-1939, https://doi.org/10.1126/science.1123604.
Li, F. et al. (2011), “A new gene for hybrid sterility from a cross between Oryza sativa and O. glaberrima”, Plant Breeding, Vol. 130, pp. 165-171, https://doi.org/10.1111/j.1439-0523.2010.01845.x.
Li, J. et al. (2016a), “Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9”, Nature Plants, Vol. 2, 16139, https://doi.org/10.1038/nplants.2016.139.
Li, L.-F. et al. (2017), “Signatures of adaptation in the weedy rice genome”, Nature Genetics, Vol. 49, pp. 811-814, https://doi.org/10.1038/ng.3825.
Li, M. et al. (2016b), “Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system”, Frontiers in Plant Science, Vol. 7, 377, https://doi.org/10.3389/fpls.2016.00377.
Li, Q. et al. (2016c), “Development of japonica photo-sensitive genic male sterile rice lines by editing carbon starved anther using CRISPR/Cas9”, Journal of Genetics and Genomics, Vol. 43, pp. 415‑419, https://doi.org/10.1016/j.jgg.2016.04.011.
Li, S., D. Yang and Y. Zhu (2007), “Characterization and use of male sterility in hybrid rice breeding”, Journal of Integrative Plant Biology, Vol. 49, pp. 791-804, https://doi.org/10.1111/j.1744-7909.2007.00513.x.
Li, S. et al. (2019), “Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing”, Journal of Integrative Plant Biology, Vol. 61, pp. 1201-1205, https://doi.org/10.1111/jipb.12774.
Li, T. et al. (2016d), “TALEN-mediated homologous recombination produces site-directed DNA base change and herbicide-resistant rice”, Journal of Genetics and Genomics, Vol. 43, pp. 297-305, https://doi.org/10.1016/j.jgg.2016.03.005.
Li, T. et al. (2012), “High-efficiency TALEN-based gene editing produces disease-resistant rice”, Nature Biotechnology, Vol. 30, pp. 390-392, https://doi.org/10.1038/nbt.2199.
Li, X. et al. (2018), “Expanding the scope of CRISPR/Cpf1-mediated genome editing in rice”, Molecular Plant, Vol. 11, pp. 995-998, https://doi.org/10.1016/j.molp.2018.03.009.
Li, X. et al. (2017), “High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing”, Journal of Genetics and Genomics, Vol. 44, pp. 175-178, https://doi.org/10.1016/j.jgg.2017.02.001.
Li, X. et al. (2015), “Combinations of Hd2 and Hd4 genes determine rice adaptability to Heilongjiang province, northern limit of China”, Journal of Integrative Plant Biology, Vol. 57, pp. 698-707, https://doi.org/10.1111/jipb.12326.
Li, Z., A. Hayashimoto and N. Murai (1992), “A sulfonylurea herbicide resistance gene from Arabidopsis thaliana as a new selectable marker for production of fertile transgenic rice plants”, Plant Physiology, Vol. 100, pp. 662-668, https://www.jstor.org/stable/4274685.
Lin, S.C. and L.P. Yuan (1980), “Hybrid rice breeding in China”, in Innovative Approaches in Rice Breeding, International Rice Research Institute, pp. 35-51.
Linares, O.F. (2002), “African rice (Oryza glaberrima): History and future potential”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 99, pp. 16360–16365, https://doi.org/10.1073/pnas.252604599.
Ling, Y., L. Ang and Z. Weilin (2019), “Current understanding of the molecular players involved in resistance to rice planthoppers”, Pest Management Science, Vol. 75, pp. 2566-2574, https://doi.org/10.1002/ps.5487.
Liu, J. et al. (2017a), “GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice”, Nature Plants, Vol. 10, 17043, https://doi.org/10.1038/nplants.2017.43.
Liu, J. et al. (2017b), “Methane emissions and microbial communities as influenced by dual cropping of Azolla along with early rice”, Scientific Reports, Vol. 7, 40635, https://doi.org/10.1038/srep40635.
Liu, W. et al. (2013), “Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae”, Molecular Plant, Vol. 6, pp. 605-620, https://doi.org/10.1093/mp/sst015.
Liu, Y. et al. (2015), “A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice”, Nature Biotechnology, Vol. 33, pp. 301-305, https://doi.org/10.1038/nbt.3069.
Long, Y. et al. (2008), “Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 105, pp. 18871-18876, https://doi.org/10.1073/pnas.0810108105.
Lou, D. et al. (2017), “OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice”, Frontiers in Plant Science, Vol. 8, pp. 993-1007, https://doi.org/10.3389/fpls.2017.00993.
Lu, B.R., X. Yang and N.C. Ellstrand (2016), “Fitness correlates of crop transgene flow into weedy populations: a case study of weedy rice in China and other examples”, Evolutionary Applications, Vol. 9, p. 857-870, https://doi.org/10.1111/eva.12377.
Lu, H.P. et al. (2018), “Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis”, Nature Plants, Vol. 4, pp. 338-344, https://doi.org/10.1038/s41477-018-0152-7.
Lu, Y. and J.-K. Zhu (2017), “Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system”, Molecular Plant, Vol. 10, p. 523-525, https://doi.org/10.1016/j.molp.2016.11.013.
Luo, D. et al. (2013), “A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice”, Nature Genetics, Vol. 45, pp. 573-577, https://doi.org/10.1038/ng.2570.
Luo, J. et al. (2013), “An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice”, The Plant Cell, Vol. 25, pp. 3360-3376, https://www.jstor.org/stable/23598355.
Lv, S. et al. (2018), “Genetic control of seed shattering during African rice domestication”, Nature Plants, Vol. 4, pp. 331-337, https://doi.org/10.1038/s41477-018-0164-3.
Ma, L. et al. (2015), “TALEN-based mutagenesis of lipoxygenase LOX3 enhances the storage tolerance of rice (Oryza sativa) seeds”, PLoS ONE., Vol. 10, e0143877, https://doi.org/10.1371/journal.pone.0143877.
Macías, F.A. et al. (2006), “Bioactive steroids from Oryza sativa L”, Steroids, Vol. 71, pp. 603-608, https://doi.org/10.1016/j.steroids.2006.03.001.
Macovei, A. et al. (2018), “Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus”, Plant Biotechnology Journal, Vol. 16, pp. 1918-1927, https://doi.org/10.1111/pbi.12927.
Maeda, H. et al. (2019), “A rice gene that confers broad-spectrum resistance to β-triketone herbicides”, Science, Vol. 365, pp. 393-396, https://doi.org/10.1126/science.aax0379.
Mao, H. et al. (2010), “Linking differential domain functions of the GS3 protein to natural variation of grain size in rice”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 107, pp. 19579-19584, https://doi.org/10.1073/pnas.1014419107.
Martínez, C.P. et al. (1994), “Nuclear DNA content of ten rice species as determined by flow cytometry”, The Japanese Journal of Genetics, Vol. 69, pp. 513-523, https://doi.org/10.1266/jjg.69.513.
Matsubara, K. et al. (2012), “Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering”, Plant and Cell Physiology, Vol. 53, pp. 709-716, https://doi.org/10.1093/pcp/pcs028.
Matsumura, M. and S. Sanada-Morimura (2010), “Recent status of insecticide resistance in asian rice planthoppers”, Japan Agricultural Research Quarterly, Vol. 44, pp. 225-230, https://doi.org/10.6090/jarq.44.225.
Matsumura, M. et al. (2008), “Species-specific insecticide resistance to imidacloprid and fipronil in the rice planthoppers Nilaparvata lugens and Sogatella furcifera in East and South-east Asia”, Pest Management Science, Vol. 64, pp. 1115–1121, https://doi.org/10.1002/ps.1641.
Matsuo, T. (1952), “Genecological studies on cultivated rice”, Bulletin of the National Institute of Agricultural Sciences Series D3, pp. 1-111 (in Japanese).
Mbodj, D. et al. (2018), “Arbuscular mycorrhizal symbiosis in rice: Establishment, environmental control and impact on plant growth and resistance to abiotic stresses”, Rhizosphere, Vol. 8, pp. 12-26, https://doi.org/10.1016/j.rhisph.2018.08.003.
McCouch, S.R. et al. (2002), “Development and mapping of 2 240 new SSR markers for rice (Oryza sativa L.)”, DNA Research, Vol. 9, pp. 199-207, https://doi.org/10.1093/dnares/9.6.199.
Mentlak, T.A. et al. (2012), “Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease”, The Plant Cell, Vol. 24, pp. 322-335, https://doi.org/10.1105/tpc.111.092957.
Messeguer, J. et al. (2004), “A field study of pollen-mediated gene flow from Mediterranean GM rice to conventional rice and the red rice weed”, Molecular Breeding, Vol. 13, pp. 103-112, https://doi.org/10.1023/B:MOLB.0000012285.39859.9d.
Miao, C. et al. (2018), “Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity”, Proceeding of the National Academy of Sciences of the United States of America, Vol. 115, pp. 6058-6063, https://doi.org/10.1073/pnas.1804774115.
Mishra, R., R.K. Joshi and K. Zhao (2018), “Genome editing in rice: Recent advances, challenges, and future implications”, Frontiers in Plant Science, Vol. 9, 1361, https://doi.org/10.3389/fpls.2018.01361.
Mithöfer, A., W. Boland and M.E. Maffei (2009), “Chemical ecology of plant-insect interactions”, Annual Plant Reviews Book Series, Vol. 34, pp. 261-291, https://doi.org/10.1002/9781119312994.apr0369.
Miura, K. et al. (2010), “OsSPL14 promotes panicle branching and higher grain productivity in rice”, Nature Genetics, Vol. 42, pp. 545-549, https://doi.org/10.1038/ng.592.
Miyabayashi, T. et al. (2007), “Genome size of twenty wild species of Oryza Species determined by flow cytometric and chromosome analyses”, Breeding Science, Vol. 57, pp. 73-78, https://doi.org/10.1270/jsbbs.57.73.
Mizuta, Y., Y. Harushima and N. Kurata (2010), “Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 107, pp. 20417-20422, https://doi.org/10.1073/pnas.1003124107.
Moldenhauer, K.A.K and J.H. Gibbons (2003), “Rice morphology and development”, in C.W. Smitha and R.H. Didlay (eds.), Rice: Origin, History, Technology, and Production, John Wiley and Sons, New York, pp. 103-127.
Moody, K. (1989), Weeds, Reported in Rice in South and Southeast Asia, International Rice Research Institute, Philippines.
Morales, F.J. (2008), “Cereal viruses: Rice”, in B.W.J. Mahy and M.H.V. van Regenmortel (eds.), Encyclopedia of Virology, 3rd edition, pp. 482-489, https://doi.org/10.1016/B978-012374410-4.00699-3.
Morinaga, T. (1968), “Origin and geographic distribution of Japanese rice”, Tropical Agriculture Research Series, Vol. 3, pp. 1-15.
Morishima, H. and H.-I. Oka (1960), “The pattern of interspecific variation in the genus Oryza: Its quantitative representation by statistical methods”, Evolution, Vol. 14, pp. 153-165, https://doi.org/10.1111/j.1558-5646.1960.tb03074.x.
Morishima, H., K. Hinata and H.-I. Oka (1963), “Comparison of modes of evolution of cultivated forms from two wild rice species, Oryza breviligulata and O. perennis”, Evolution, Vol. 17, pp. 70-181, https://doi.org/10.1270/jsbbs1951.12.153.
Morishima, H., K. Hinata and H.-I. Oka (1962), “Comparison between two cultivated rice species, Oryza sativa L. and O. glaberrima Steud.”, Japanese Journal of Breeding Science, Vol. 12, pp. 153-165.
Multani, D.S. et al. (2003), “Alien genes introgression and development of monosomic alien additional lines from Oryza latifolia Desv. to rice, Oryza sativa L.”, Theoretical and Applied Genetics, Vol. 107, p. 395-405, https://doi.org/10.1007/s00122-003-1214-3.
Multani, D.S. et al. (1994), “Development of monosomic alien addition lines and introgression of genes from Oryza australiensis Domin. to cultivated rice O. sativa L.”, Theoretical and Applied Genetics, Vol. 88, pp. 102-109, https://doi.org/10.1007/BF00222401.
Muniyappa, V. and S.P. Raychaudhuri (1988), “Rice yellow dwarf disease”, in K. Maramorosch and S.P. Raychaudhuri (eds.), Mycoplasma Diseases of Crops, Springer, pp. 233-284, https://doi.org/10.1007/978-1-4612-3808-9_14.
Nadir, S. et al. (2019), “A novel discovery of a long terminal repeat retrotransposon-induced hybrid weakness in rice”, Journal of Experimental Botany, Vol. 70, pp. 1197-1207, https://doi.org/10.1093/jxb/ery442.
Nagai, I. (1959), “Procedure”, in Japonica Rice – Its Breeding and Culture, Yokendo, Tokyo, pp. 408-435.
Nagao, S. and T. Takano (1938), “Duration of fertilizing capacity of pollen and stigma in rice”, in S. Nagano (ed.), Commemoration Papers in honour of Prof M. Akemine, Yokendo, Tokyo, pp. 88-92 (in Japanese).
Nakagawa, H. and H. Kato (2017), “Induced mutations for food and energy security: Challenge of inducing unique mutants for new cultivars and molecular research”, NARO Bulletin, Crop Science Institute, Vol. 1, pp. 33-124.
Nakamura, T. (2003), “A method for discriminating the nymphs of two species of predacious mirid bug, Cyrtorhinus lividipennis Reuter and Tytthus chinensis (Stål), and their occurrence in paddy fields”, Kyushu Plant Protection Research, Vol. 49, pp. 77-82, https://jglobal.jst.go.jp/en/detail?JGLOBAL_ID=200902205564728147 (in Japanese with English abstract).
Nakashima, K. and T. Hayashi (1995), “Multiplication and distribution of rice yellow dwarf phytoplasma in infected tissues of rice and green rice leafhopper nephotettix cincticeps”, Japanese Journal of Phytopathology, Vol. 61, pp. 451-455, https://doi.org/10.3186/jjphytopath.61.451.
Nakayama, R. (1934), “The artificial germination of rice pollen”, Agriculture and Horticulture, Vol. 9, pp. 1917-1926 (in Japanese).
Nandi, S. et al. (2002), “Expression of human lactoferrin in transgenic rice grains for the application in infant formula”, Plant Science, Vol. 163, pp. 713-722, https://doi.org/10.1016/S0168-9452(02)00165-6.
NARO (accessed 22 March 2022), Genebank Project, National Agriculture and Food Research Organization, Japan, https://www.gene.affrc.go.jp/databases.php.
Nemoto, Y. et al. (2016), “Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7”, The Plant Journal, Vol. 86, pp. 221-233, https://doi.org/10.1111/tpj.13168.
Nezu, M., T.C. Katayama and H. Kihara (1960), “Genetic study of the genus Oryza. Crossability and chromosome affinity among 17 species”, Seiken Zihô (Reports of Kihara Institute for Biological Researches), Vol. 11, pp. 1-11.
Nguyen, G.N. et al. (2017), “Duplication and loss of function of genes encoding RNA polymerase III subunit C4 causes hybrid incompatibility in rice”, G3: Genes, Genomes Genetics, Vol. 7, pp. 2565‑2575, https://doi.org/10.1534/g3.117.043943.
Niang, A. et al. (2017), “Variability and determinants of yields in rice production systems of West Africa”, Field Crops Research, Vol. 207, pp. 1-12, https://doi.org/10.1016/j.fcr.2017.02.014.
Nicol, J.M. et al. (2011), “Current nematode threats to world agriculture”, in J. Jones, G. Gheysen and C. Fenoll (eds.), Genomics and Molecular Genetics of Plant-Nematode Interactions, Springer, Dordrecht, pp. 21-43, https://doi.org/10.1007/978-94-007-0434-3_2.
Niizeki, H. and K. Oono (1968), “Induction of haploid rice plant from anther culture”, Proceedings of the Japan Academy, Vol. 44, pp. 554-557, https://doi.org/10.2183/pjab1945.44.554.
Niño-Liu, D.O., P.C. Ronald and A.J. Bogdanove (2006), “Xanthomonas oryzae pathovars: Model pathogens of a model crop”, Molecular plant pathology. Vol. 7, pp. 303-324, https://doi.org/10.1111/j.1364-3703.2006.00344.x.
Nishida, K. et al. (2016), “Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems”, Science, Vol. 353/6305, https://doi.org/10.1126/science.aaf8729.
Nishimasu, H. et al. (2018), “Engineered CRISPR-Cas9 nuclease with expanded targeting space”, Science, Vol. 361, pp. 1259-1262, https://doi.org/10.1126/science.aas9129.
Nishiyama, Y. (1995), “Factors and mechanisms causing cool weather damage”, in T. Matsuo et al. (eds.), Science of the Rice Plant: Physiology, Food and Agriculture Policy Research Center, Tokyo, pp. 776-793.
OECD (2016), “Revised Consensus Document on Compositional Considerations for New Varieties of Rice (Oryza sativa): Key Food and Feed Nutrients, Anti-nutrients and Other Constituents”, Series on the Safety of Novel Foods and Feeds, No. 28, OECD Publishing, Paris, http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/jm/mono(2016)38&doclanguage=en.
Ohno, H. et al. (2018), “Longer mesocotyl contributes to quick seedling establishment, improved root anchorage, and early vigor of deep-sown rice”, Field Crops Research, Vol. 228, pp. 84–92, https://doi.org/10.1016/j.fcr.2018.08.015.
Oikawa, T. et al. (2015), “The birth of a black rice gene and its local spread by introgression”, The Plant Cell, Vol. 27, pp. 2401-2414, https://doi.org/10.1105/tpc.15.00310.
Oka, H. (1954), “Varietal variation of the responses to day-length and temperature and the number of the days to growth period”, Japanese Journal of Breeding, Vol. 4, pp. 92-100, https://doi.org/10.1270/jsbbs1951.4.92 (in Japanese with English abstract).
Oka, H.-I. (1992), “Ecology of wild rice planted in Taiwan III. Differences in regenerating strategies among genetic stocks”, Botanical Bulletin of Academia Sinica, Vol. 33, pp. 133-140.
Oka, H.-I. (1988a), “Ancestors of cultivated rice”, in Origin of Cultivated Rice, Japan Scientific Societies Press, Tokyo/Elsevier, Amsterdam, pp. 18-22.
Oka, H.-I. (1988b), “Variations in the breeding systems”, in Origin of Cultivated Rice, Japan Scientific Societies Press, Tokyo/Elsevier, Amsterdam, pp. 32-36.
Oka, H.-I. (1988c), “The dynamics of domestication”, in Origin of Cultivated Rice, Japan Scientific Societies Press Tokyo/Elsevier Amsterdam, pp. 114-123.
Oka, H.-I. (1957), “Genic analysis for the sterility of hybrids between distantly related varieties of cultivated rice”, Journal of Genetics, Vol. 55, pp. 397-409, https://doi.org/10.1007/bf02984059.
Oka, H.-I. and H. Morishima (1967), “Variations in the breeding systems of a wild rice, Oryza perennis”, Evolution, Vol. 21, pp. 249-258, https://doi.org/10.1111/j.1558-5646.1967.tb00153.x.
Oka, H.-I. and W.T. Chang (1961), “Hybrid swarms between wild and cultivated rice species, Oryza perennis and O. sativa”, Evolution, Vol. 15, pp. 418-430, https://doi.org/10.2307/2406310.
Okazaki, M. et al. (2013), “Whole mitochondrial genome sequencing and transcriptional analysis to uncover an RT102-type cytoplasmic male sterility-associated candidate gene derived from Oryza rufipogon”, Plant and Cell Physiology, Vol. 54, pp. 1560-1568, https://doi.org/10.1093/pcp/pct102.
Okuno, K and K. Ebana (2003), “Identification of QTL controlling allelopathic effects in rice: Genetic approaches to biological control of weeds”, Japan Agricultural Research Quarterly, Vol. 37, p. 77-81, https://doi.org/10.6090/jarq.37.77.
Olofsdotter, M., D. Navarez and K. Moody (1995), “Allelopathic potential in rice (Oryza sativa L.) germplasm”, Annals of Applied Biology, Vol. 127, pp. 543-560, https://doi.org/10.1111/j.1744-7348.1995.tb07611.x.
Olofsdotter, M., D. Navarez and M. Rebulanan (1997), “Rice allelopathy - Where are we and how far can we get?”, Brighton Crop Protection Conference: Weeds, Vol. 1, pp. 99-104.
Olofsdotter, M.et al. (2002), “Why phenolic acids are unlikely primary allelochemicals in rice”, Journal of Chemical Ecology, Vol. 28, pp. 229-242, https://doi.org/10.1023/A:1013531306670.
Olofsdotter, M.et al. (1999), “Weed suppressing rice cultivars - Does allelopathy play a role?”, Weed Research, Vol. 39, pp. 441-454, https://doi.org/10.1046/j.1365-3180.1999.00159.x.
Oryzabase (n.d.), website, https://shigen.nig.ac.jp/rice/oryzabase/
Ou, S.H. (1985), Rice Diseases, 2nd edition, Commonwealth Mycological Institute, p. 380.
Ouyang, S. et al. (2007), “The TIGR Rice Genome Annotation Resource: Improvements and new features”, Nucleic Acids Research, Vol. 35, pp. 883-887, https://doi.org/10.1093/nar/gkl976.
Padma, V. and B.M. Reddy (2000), “Evaluation of rice genotypes for dormancy duration and seed storability under natural and accelerated ageing”, Seed Research, Vol. 28, pp. 158-165.
Paine, J. et al. (2005), “Improving the nutritional value of Golden Rice through increased pro-vitamin A content”, Nature Biotechnology, Vol. 23, pp. 482-487, https://doi.org/10.1038/nbt1082.
Pandey, S. and L. Velasco (2005), “Trends in crop establishment methods in Asia and research issues”, in K. Toriyama, K.L. Heong and B. Hardy (eds.), Rice is Life: Scientific Perspectives for the 21st Century, IRRI and JIRCAS, pp. 178-181.
Pandit, P.S. et al. (2016), “Deciphering community structure of methanotrophs dwelling in rice rhizospheres of an Indian rice field using cultivation and cultivation-independent approaches”, Microbial Ecology, Vol. 71, pp. 634-644, https://doi.org/10.1007/s00248-015-0697-1.
Pang, Y. et al. (2017), “Recurrent selection breeding by dominant male sterility for multiple abiotic stresses tolerant rice cultivars”, Euphytica, Vol. 213, Article 268, https://doi.org/10.1007/s10681-017-2055-5.
Park, C.H. et al. (2012), “The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern–triggered immunity in rice”, The Plant Cell, Vol. 24, pp. 4748-4762, https://doi.org/10.1105/tpc.112.105429.
Pathak, M.D. and Z.R. Khan (1994), Insect Pests of Rice, International Rice Research Institute, Manila, Philippines, https://www.cabi.org/isc/abstract/19951100418.
Pathak, M.D. and G. Khush (1979), “Studies of varietal resistance in rice to the brown planthopper at the International Rice Research Institute”, in International Rice Research Institute (ed.), Brown Planthopper: Threat to Rice Production in Asia, International Rice Research Institute, Manila, Philippines, pp. 285-301.
Peng, Y., R. van Wersch, Y. Zhang (2018), “Convergent and divergent signalling in PAMP-triggered immunity and effector-triggered immunity”, Molecular Plant-Microbe Interactions Journal, Vol. 31, pp. 403-409, https://doi.org/10.1094/MPMI-06-17-0145-CR.
Podevin, N. et al. (2013), “Site-directed nucleases: A paradigm shift in predictable, knowledge-based plant breeding”, Trends in Biotechnology, Vol. 31, pp. 375-383, https://doi.org/10.1016/j.tibtech.2013.03.004.
Pu, D.-q. et al. (2014), “Flower-visiting insects and their potential impact on transgene flow in rice”, Journal of Applied Ecology, Vol. 51, pp. 1357-1365, www.jstor.org/stable/24032573.
Pusadee, T. et al. (2013), “Population structure of the primary gene pool of Oryza sativa in Thailand”, Genetic Resources and Crop Evolution, Vol. 60, pp. 335-353, https://doi.org/10.1007/s10722-012-9839-1.
Rao, A.N., N. Chandrasena and H. Matsumoto (2017), “Rice weed management in the Asian-Pacific region: An overview”, in A.N. Rao and H. Matsumoto (eds.), Weed Management in Rice in the Asian-Pacific Region, Asian-Pacific Weed Science Society, Hyderabad, pp.1-41, http://oar.icrisat.org/10210/.
Rao, N.K. and M.T. Jackson (1997), “Variation in seed longevity of rice cultivars belonging to different isozyme groups”, Genetic Resources and Crop Evolution, Vol. 44, pp. 159-164, https://doi.org/10.1023/A:1008642318474.
Rao, N.K. and M.T. Jackson (1996a), “Seed longevity of rice cultivars and strategies for their conservation in genebanks”, Annals of Botan, Vol. 77, pp. 251-260, https://doi.org/10.1006/anbo.1996.0029.
Rao, N.K and M.T. Jackson (1996b), “Effect of sowing date and harvest time on longevity of rice seeds”, Seed Science Research, Vol. 7, pp. 13-20, https://doi.org/10.1017/S0960258500003317.
Rao N.K. and M.T. Jackson (1996c), “Seed production environment and storage longevity of japonica rices (Oryza sativa L)”, Seed Science Research, Vol. 6, pp. 17-21, https://doi.org/10.1017/S0960258500002956.
Ren, G. et al. (2005), “A new gamete eliminator from Oryza glaberrima”, Rice Genetics Newsletter, Vol.22, pp.45-47.
Ren, J. et al. (2016), “Bph32, a novel gene encoding an unknown SCR domain-containing protein, confers resistance against the brown planthopper in rice”, Scientific Reports, Vol. 6, 37645, https://doi.org/10.1038/srep37645.
Ressing, W.H. et al. (1986), Illustrated Guide to Integrated Pest Management in Rice in Tropical Asia, International Rice Research Institute, p. 411.
Reuscher, S. et al. (2018), “Assembling the genome of the African wild rice Oryza longistaminata by exploiting synteny in closely related Oryza species”, Communications Biology, Vol. 1, Article 162, https://doi.org/10.1038/s42003-018-0171-y.
Roberts, E.H. (1961), “The viability of rice seed in relation to temperature, moisture content, and gaseous environment”, Annals of Botany, Vol. 25, pp. 381-390, https://www.jstor.org/stable/42907599.
Rodenburg, J. et al. (2019), “Status quo of chemical weed control in rice in sub-Saharan Africa”, Food Security, Vol. 11, p. 69-92, https://doi.org/10.1007/s12571-018-0878-0.
Rodenburg, J. et al. (2017), “Genetic variation and host-parasite specificity of Striga resistance and tolerance in rice: The need for predictive breeding”, New Phytologist, Vol. 214, pp. 1267-1280, https://doi.org/10.1111/nph.14451.
Rong, J. et al. (2012), “Scale effect on rice pollen-mediated gene flow: Implications in assessing transgene flow from genetically engineered plants”, Annals of Applied Biology, Vol. 161, pp. 3-12, https://doi.org/10.1111/j.1744-7348.2012.00545.x.
Rutger, J.N. (1992), “Impact of mutation breeding in rice – A review”, Mutation Breeding Review, Vol. 8, pp. 2-23.
Sacks, E.J. (2013), “Perennial rice: Challenges and opportunities”, in Perennial Crops for Food Security: Proceedings of the FAO Expert Workshop, Food and Agriculture Organization of the United Nations, pp. 16-26.
Saito, K. et al. (2018), “Progress in varietal improvement for increasing upland rice productivity in the tropics”, Plant Production Science, Vol. 21, pp. 145-158, https://doi.org/10.1080/1343943X.2018.1459751.
Saito, K. et al. (2013), “Towards a better understanding of biophysical determinants of yield gaps and the potential for expansion of the rice area in Africa”, in M.C.S. Wopereis et al. (eds.), Realizing Africa’s Rice Promise, CABI, pp. 188-203, https://doi.org/10.1079/9781845938123.0188.
Sakai, H. et al. (2013), “Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics”, Plant and Cell Physiology, Vol. 54, e6, https://doi.org/10.1093/pcp/pcs183.
Sakai, H. et al. (2011), “Distinct evolutionary patterns of Oryza glaberrima deciphered by genome sequencing and comparative analysis”, The Plant Journal, Vol. 66, pp. 796–805, https://doi.org/10.1111/j.1365-313X.2011.04539.x.
Sano, Y. (1990), “The genic nature of gamete eliminator in rice”, Genetics, Vol. 125, pp. 183-191.
Sano, Y., Y.-E. Chu and H. Oka (1979), “Genetic studies of speciation in cultivated rice. 1. Genetic analysis for the F1 sterility between O. sativa L. and O. glaberrima Steud”, The Japanese Journal of Genetics, Vol. 54, pp. 121-132, https://doi.org/10.1266/jjg.54.121.
Sasaki, A. et al. (2002), “A mutant gibberellin-synthesis gene in rice”, Nature, Vol. 416, pp. 701-702, https://doi.org/10.1038/416701a.
Sato, Y. and S. Yokoya (2008), “Effects of male sterility caused by low temperature at the booting stage on out-crossing rates in rice (Oryza sativa L.)”, Breeding Research, Vol. 10, pp. 127-134, https://doi.org/10.1270/jsbbr.10.127 (in Japanese with English abstract).
Sato, Y. et al. (2013), “RiceXPro version 3.0: Expanding the informatics resource for rice transcriptome”, Nucleic Acids Research, Vol. 41, pp. D1206-D1213, https://doi.org/10.1093/nar/gks1125.
Sato, Y. et al. (2012), “RiceFREND: A platform for retrieving coexpressed gene networks in rice”, Nucleic Acids Research, Vol. 41, pp. D1214-D1221, https://doi.org/10.1093/nar/gks1122.
Sato, Y. et al. (2010), “RiceXPro: A platform for monitoring gene expression in japonica rice grown under natural field conditions”, Nucleic Acids Research, Vol. 39 (Suppl. 1), pp. D1141-D1148, https://doi.org/10.1093/nar/gkq1085.
Schwartz, C. et al. (2020), “CRISPR–Cas9-mediated 75.5-Mb inversion in maize”, Nature Plants, Vol. 6, pp. 1427-1431, https://doi.org/10.1038/s41477-020-00817-6.
Septiningsih, E.M. et al. (2013), “QTL mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace Ma-Zhan Red”, Theoretical and Applied Genetics, Vol. 126, pp. 1357-1366, https://doi.org/10.1007/s00122-013-2057-1.
Sessitsch, A. et al. (2015), “Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis”, Molecular Plant-Microbe Interactions, Vol. 25, pp. 28-36, https://doi.org/10.1094/MPMI-08-11-0204.
Shabir, H.W. et al. (2015), “Transgenic rice: Advancements and achievements”, Advancements in Genetic Engineering, Vol. 4, pp. 1-3, https://doi.org/10.4172/2169-0111.1000133.
Shabuer, G. et al. (2015), “Plant pathogenic anaerobic bacteria use aromatic polyketides to access aerobic territory”, Science, Vol. 350, pp. 670-674, https://doi.org/10.1126/science.aac9990.
Shan, Q. et al. (2015), “Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology”, Plant Biotechnology Journal, Vol. 13, pp. 791-800, https://doi.org/10.1111/pbi.12312.
Shao, G. et al. (2017), “CRISPR/CAS9-mediated editing of the fragrant gene Badh2 in rice”, Chinese Journal of Rice Science, Vol. 31, pp. 216-222, https://doi.org/10.16819/j.1001-7216.2017.6098 (in Chinese).
Shen, R. et al. (2017), “Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice”, Nature Communications, Vol. 8, p. 1310, https://doi.org/10.1038/s41467-017-01400-y.
Shenton, M. et al. (2016), “Effect of wild and cultivated rice genotypes on rhizosphere bacterial community composition”, Rice, Vol. 9, Article 42, https://doi.org/10.1186/s12284-016-0111-8.
Shepard, B.M., A.T. Barrion and J.A. Litsinger (1995), Rice-Feeding Insects of Tropical Asia, International Rice Research Institute, Manila, Philippines, p. 228.
Shigematsu, Y. et al. (1982), “Sterols and asparagine in the rice plant, endogenous factors related to resistance against the brown planthopper (Nilaparvata lugens)”, Agricultural and Biological Chemistry, Vol. 46, pp. 2877-2879, https://doi.org/10.1271/bbb1961.46.2877.
Shimada, T. (2002), “Daily activity pattern and habitat use of Greater White-fronted Geese wintering in Japan: Factors of the population increase”, Waterbirds, Vol. 25, pp. 371-377, https://doi.org/10.1675/1524-4695(2002)025[0371:DAPAHU]2.0.CO;2.
Shimamoto, K. et al. (1989), “Fertile transgenic rice plants regenerated from transformed protoplasts”, Nature, Vol. 338, pp. 274-276, https://doi.org/10.1038/338274a0.
Shimatani, Z. et al. (2017) “Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion”, Nature Biotechnology, Vol. 35, pp. 441-443, https://doi.org/10.1038/nbt.3833.
Shimono, M. et al. (2007), “Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance”, The Plant Cell, Vol. 19, pp. 2064-2076, https://doi.org/10.1105/tpc.106.046250.
Shivrain, V.K. et al. (2008), “Maximum outcrossing rate and genetic compatibility between red rice (Oryza sativa) biotypes and Clearfield™ rice”, Weed Science, Vol. 56, pp. 807-813, https://doi.org/10.1614/WS-08-026.1.
Shivrain, V.K. et al. (2007), “Gene flow between Clearfield™ rice and red rice”, Crop Protection, Vol. 26, pp. 349-356, https://doi.org/10.1016/j.cropro.2005.09.019.
Shomura, A. et al. (2008), “Deletion in a gene associated with grain size increased yields during rice domestication”, Nature Genetics, Vol. 40, pp. 1023-1028, https://doi.org/10.1038/ng.169.
Singh, R. et al. (2016), “Magnaporthe oryzae effector AVR-Pii helps to establish compatibility by inhibition of the rice NADP-Malic enzyme resulting in disruption of oxidative burst and host innate immunity”, Molecules and Cells, Vol. 39, pp. 426-438, https://doi.org/10.14348/molcells.2016.0094.
Singh, S. et al. (2001), “Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106”, Theoretical and Applied Genetics, Vol. 102, pp. 1011-1015, https://doi.org/10.1007/s001220000495.
Sitch, L.A. (1990), “Incompatibility barriers operating in crosses of Oryza sativa with related species and genera”, in J.P. Gustafson (ed.), Gene Manipulation in Plant Improvement II, Plenum Press, New York, pp. 77-93, https://doi.org/10.1007/978-1-4684-7047-5_5.
Sleper, D.A. and J.M. Poehlman (2006), “Breeding rice”, in Breeding Field Crops, 5th edition, Blackwell Publishing, Ames, pp. 239-257.
Sōgawa, K. (1982), “The rice brown planthopper: Feeding physiology and host plant interactions”, Annual Review of Entomology, Vol. 27, pp. 49-73, https://doi.org/10.1146/annurev.en.27.010182.000405.
Song, B.K. et al. (2014), “Malaysian weedy rice shows its true stripes: Wild Oryza and elite rice cultivars shape agricultural weed evolution in Southeast Asia”, Molecular Ecology, Vol. 23, pp. 5003-5017, https://doi.org/10.1111/mec.12922.
Song, Z.P. et al. (2003), “Gene flow from cultivated rice to the wild species Oryza rufipogon under experimental field conditions”, New Phytologist, Vol. 157, pp. 657-665, https://doi.org/10.1046/j.1469-8137.2003.00699.x.
Stein, J.C. et al. (2018), “Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza”, Nature Genetics, Vol. 50, pp. 285‑296, https://doi.org/10.1038/s41588-018-0040-0.
Stevenson, P.C. et al. (1996), “Schaftosides from rice phloem as feeding inhibitors and resistance factors to brown planthoppers, Nilaparvata lugens”, Proceedings of the 9th International Symposium on Insect-Plant Relationships, Vol. 53, pp. 246-249, https://doi.org/10.1007/978-94-009-1720-0_56.
Sudianto, E. et al. (2013), “ClearfieldⓇ rice: Its development, success, and key challenges on a global perspective”, Crop Protection, Vol. 49, pp. 40-51, https://doi.org/10.1016/j.cropro.2013.02.013.
Sugita, K. et al. (2005), “Genetically modified rice seeds accumulating GLP-1 analogue stimulate insulin secretion from a mouse pancreatic beta-cell line”, FEBS Letters, Vol. 579, pp. 1085-1088, https://doi.org/10.1016/j.febslet.2004.12.082.
Suh, H.S., Y.I. Sato and H. Morishima (1997), “Genetic characterization of weedy rice (Oryza sativa L.) based on morpho-physiology, isozymes and RAPD markers”, Theoretical and Applied Genetics, Vol. 94, pp. 316-321, https://doi.org/10.1007/s001220050417.
Suketomo, C., T. Kazama and K. Toriyama (2020), “Fertility restoration of Chinese wild rice-type cytoplasmic male sterility by CRISPR/Cas9-mediated genome editing of nuclear-encoded RETROGRADE-REGULATED MALE STERILITY”, Plant Biotechnology, Vol. 37, pp. 285-292, https://doi.org/10.5511/plantbiotechnology.20.0326b.
Sun, Y. et al. (2017), “Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes”, Frontiers in Plant Science, Vol. 8, 298, https://doi.org/10.3389/fpls.2017.00298.
Sun, C. et al. (2016a), “RPAN: Rice pan-genome browser for ∼3 000 rice genomes”, Nucleic Acids Research, Vol. 45. pp. 597-605, https://doi.org/10.1093/nar/gkw958.
Sun, Y. et al. (2016b), “Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase”, Molecular Plant, Vol. 9, pp. 628-631, https://doi.org/10.1016/j.molp.2016.01.001.
Suzuki, K. et al. (2011), “Prevention of allergic asthma by vaccination with transgenic rice seed expressing mite allergen: Induction of allergen-specific oral tolerance without bystander suppression”, Plant Biotechnology Journal, Vol. 9, pp. 982-990, https://doi.org/10.1111/j.1467-7652.2011.00613.x.
Suzuki, Y. (2003), Seed Biology, Tohoku-University Press, Sendai, p. 204 (in Japanese).
Sweeney, M.T. et al. (2007), “Global dissemination of a single mutation conferring white pericarp in rice”, PLoS Genetics, Vol. 3, e133, https://doi.org/10.1371/journal.pgen.0030133.
Taguchi, K., K. Doi and A. Yoshimura (1999), “RFLP mapping of S19, a gene for F1 pollen semi-sterility found in backcross progeny of Oryza sativa and O. glaberrima”, Rice Genetics Newsletter, Vol. 16, p. 70-71, https://archive.gramene.org/newsletters/rice_genetics/rgn16/v16p70.html.
Takagi, H. et al. (2005), “Oral immunotherapy against a pollen allergy using a seed-based peptide vaccine”, Plant Biotechnology Journal, Vol. 3, pp. 521-533, https://doi.org/10.1111/j.1467-7652.2005.00143.x.
Takahashi, N. and Y. Suzuki (1975), “Factor affecting the viability of seed with special references to maturity and longevity”, Japanese Committee for the International Biological Program synthesis, Vol. 5, pp. 63-69.
Takaiwa, F. et al. (2015), “Rice seed for delivery of vaccines to gut mucosal immune tissues”, Plant Biotechnology Journal, Vol. 13, pp. 1041-1055, https://doi.org/10.1111/pbi.12423.
Takatsuji, H. (2014), “Development of disease-resistant rice using regulatory components of induced disease resistance”, Frontiers in Plant Science, Vol. 5, 630, https://doi.org/10.3389/fpls.2014.00630.
Tamura, Y et al. (2014), “Map-based cloning and characterization of a brown planthopper resistance gene BPH26 from Oryza sativa L. ssp. indica cultivar ADR52”, Scientific Reports, Vol. 4, Article 5872, https://doi.org/10.1038/srep05872.
Tan, L. et al. (2008), “Control of a key transition from prostrate to erect growth in rice domestication”, Nature Genetics, Vol. 40, pp. 1360-1364, https://doi.org/10.1038/ng.197.
Tan, S. et al. (2005) “Imidazolinone-tolerant crops: History, current status and future”, Pest Management Science, Vol. 61, pp. 246-257, https://doi.org/10.1002/ps.993.
Tanaka, K. and M. Matsumura (2000), “Development of virulence to resistant rice varieties in the brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae), immigrating into Japan”, Applied Entomology and Zoology, Vol. 35, pp. 529-533, https://doi.org/10.1303/aez.2000.529.
Tang, H. et al. (2017), “Multi-step formation, evolution, and functionalization of new cytoplasmic male sterility genes in the plant mitochondrial genomes”, Cell Research, Vol. 27, pp. 130-146, https://doi.org/10.1038/cr.2016.115.
Tang, H. et al. (2014), “The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts”, Molecular Plant, Vol. 7, pp. 1497-1500, https://doi.org/10.1093/mp/ssu047.
Tang, L. et al. (2017), “Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd‑accumulating indica rice without compromising yield”, Scientific Reports, Vol. 7, Article 14438, https://doi.org/10.1038/s41598-017-14832-9.
Tang, L.H. and H. Morishima (1998), “Characteristics of weed rice strains”, Rice Genetics Newsletter, Vol. 5, pp. 70-72, https://archive.gramene.org/newsletters/rice_genetics/rgn5/v5II12.html.
Tanno, H. et al. (2011), “Relation between out-crossing rate and isolation distance under cool temperature conditions at the booting stage that causes the occurrence of male sterility in rice”, Japanese Journal of Crop Science, Vol. 80, pp. 49-58, https://doi.org/10.1626/jcs.80.49 (in Japanese).
Toki, S. et al. (2006), “Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice”, The Plant Journal, Vol. 47, pp. 969-976, https://doi.org/10.1111/j.1365-313X.2006.02836.x.
Toriyama, K. et al. (1988), “Transgenic rice plants after direct gene transfer into protoplasts”, Nature Biotechnology, Vol. 6, pp. 1072-1074, https://doi.org/10.1038/nbt0988-1072.
Tozawa, Y. et al. (2001), “Characterization of rice anthranilate synthase α-subunit genes OASA1 and OASA2. Tryptophan accumulation in transgenic rice expressing a feedback-insensitive mutant of OASA1”, Plant Physiology, Vol. 126, pp. 1493-1506, https://doi.org/10.1104/pp.126.4.1493.
Tsushima, S. (1996), “Epidemiology of bacterial grain rot of rice caused by pseudomonas glumae”, Japan Agricultural Research Quarterly, Vol. 30, pp. 85-89, https://www.semanticscholar.org/paper/Epidemiology-of-Bacterial-Grain-Rot-of-Rice-Caused-Tsushima/1e7032400ed6702ea1dafb5cd0e85244d7e18bc2.
Uga, Y. et al. (2003), “Variations of floral traits in Asian cultivated rice (Oryza sativa L.) and its wild relatives (O. rufipogon Griff.)”, Breeding Science, Vol. 53, pp. 345-352, https://doi.org/10.1270/jsbbs.53.345.
Ura, H. et al. (2006), “Burkholderia gladioli associated with symptoms of bacterial grain rot and leaf-sheath browning of rice plants”, Journal of General Plant Pathology, Vol. 72, pp. 98-103, https://doi.org/10.1007/s10327-005-0256-6.
Vacher, C. et al. (2016), “The phyllosphere: Microbial jungle at the plant-climate interface”, Annual Review of Ecology, Evolution, and Systematics, Vol. 47, pp. 1-24, https://doi.org/10.1146/annurev-ecolsys-121415-032238.
Valarmathi, P. et al. (2013), “First report of Rice orange leaf disease phytoplasma (16 SrI) in rice (Oryza sativa) in India”, Australasian Plant Disease Notes, Vol. 8, pp. 141-143, https://doi.org/10.1007/s13314-013-0117-7.
Vasconcelos, M. et al. (2003), “Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene”, Plant Science, Vol. 164, pp. 371-378, https://doi.org/10.1016/S0168-9452(02)00421-1.
Vidotto, F. and A. Ferrero (2000), “Germination behaviour of red rice (Oryza sativa L.) seeds in field and laboratory conditions”, Agronomie, Vol. 20, pp. 375-382, https://doi.org/10.1051/agro:2000134.
Virmani, S.S. (1994), “Natural outcrossing mechanisms in rice”, in Heterosis and Hybrid Breeding, Monographs on Theoretical and Applied Genetics, Vol. 22, pp. 82-96, https://doi.org/10.1007/978-3-642-85115-5_3.
Wakasa, Y. and F. Takaiwa (2013), “The use of rice seeds to produce human pharmaceuticals for oral therapy”, Biotechnology Journal, Vol. 8, pp. 1133-1143, https://doi.org/10.1002/biot.201300065.
Wakasa, Y. et al. (2011), “The hypocholesterolemic activity of transgenic rice seed accumulating lactostatin, a bioactive peptide derived from bovine milk β-lactoglobulin”, Journal of Agricultural and Food Chemistry, Vol. 59, pp. 3845-3850, https://doi.org/10.1021/jf200044j.
Wang, F. et al. (2016), “Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922”, PLoS ONE, Vol. 11, e0154027, https://doi.org/10.1371/journal.pone.0154027.
Wang, K. et al. (2013a), “ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice”, New Phytologist, Vol. 198, pp. 408-418, https://doi.org/10.1111/nph.12180.
Wang, K. et al. (2013b), “Gene, protein, and network of male sterility in rice”, Frontiers in Plant Science, Vol. 4, 92, https://doi.org/10.3389/fpls.2013.00092.
Wang, L. et al. (2010), “A dynamic gene expression atlas covering the entire life cycle of rice”, The Plant Journal, Vol. 61, pp. 752-766, https://doi.org/10.1111/j.1365-313X.2009.04100.x.
Wang, M. et al. (2014), “The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication”, Nature Genetics, Vol. 46, pp. 982-988, https://doi.org/10.1038/ng.3044.
Wang, Q. et al. (2019), "Research progress on effects of straw returning on nitrogen cycling microbes and functional genes in paddy soil", Acta Agriculturae Zhejiangensis, Vol. 31, pp. 333-342, https://doi.org/10.3969/j.issn.1004-1524.2019.02.20 (in Chinese with English abstract).
Wang, W. et al. (2018), “Genomic variation in 3,010 diverse accessions of Asian cultivated rice”, Nature, Vol. 557, pp. 43-49, https://doi.org/10.1038/s41586-018-0063-9.
Wang, Y. et al. (2015), “Map-based cloning and characterization of BPH29, a B3 domain-containing recessive gene conferring brown planthopper resistance in rice”, Journal of Experimental Botany, Vol. 66, pp. 6035-6045, https://doi.org/10.1093/jxb/erv318.
Wang, Z. et al. (2006), “Cytoplasmic male sterility of rice with Boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing”, The Plant Cell, Vol. 18, pp. 676-687, https://doi.org/10.1105/tpc.105.038240.
Watanabe, Y. (1997), “Phylogeny and geographical distribution of genus Oryza”, in T. Matsuo et al. (eds.), Science of the Rice Plant: Genetics, Vol. 3, Genetics, Food and Agriculture Policy Research Center, Tokyo, pp. 29-39.
Watanabe, Y. (1993), “Classification and morphological characters of plants in genus Oryza”, in T. Matsuo and K. Hoshikawa (eds.), Science of the Rice Plant, Vol. 1. Morphology, Food and Agricultural Policy Research Center, Tokyo, pp. 23-30.
Watanarojanaporn, N. et al. (2013), “Effect of rice cultivation systems on indigenous arbuscular mycorrhizal fungal community structure”, Microbes and Environments, Vol. 28, pp. 316-324, https://doi.org/10.1264/jsme2.ME13011.
Wei, F.J. et al. (2016), “Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice”, Plant Science, Vol. 242, pp. 187-194, https://doi.org/10.1016/j.plantsci.2015.09.005.
Weng, J. et al. (2008), “Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight”, Cell Research, Vol. 18, pp. 1199-1209, https://doi.org/10.1038/cr.2008.307.
Win, K.T. et al. (2009), “Identification of two loci causing F1 pollen sterility in inter- and intraspecific crosses of rice”, Breeding Science, Vol. 59, pp. 411-418, https://doi.org/10.1270/jsbbs.59.411.
Wing, R., M.D. Purugganan and Q. Zhang (2018), “The rice genome revolution: From an ancient grain to Green Super Rice”, Nature Reviews Genetics, Vol. 19, pp. 505-517, https://doi.org/10.1038/s41576-018-0024-z.
Wu, N. et al. (2020), “Rice black-streaked dwarf virus: From multiparty interactions among plant–virus–vector to intermittent epidemics”, Molecular Plant Pathology, Vol. 21, pp. 1007-1019, https://doi.org/10.1111/mpp.12946.
Wu, W. et al. (2017), “A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication”, Nature Plants, Vol. 3, Article 17064, https://doi.org/10.1038/nplants.2017.64.
Wu, X.R., Z.H. Chen and W.R. Folk (2003), “Enrichment of cereal protein lysine content by altered tRNAlys coding during protein synthesis”, Plant Biotechnology Journal, Vol. 1, pp. 187-194, https://doi.org/10.1046/j.1467-7652.2003.00017.x.
Wu, Z. et al. (2018), “De novo genome assembly of Oryza granulata reveals rapid genome expansion and adaptive evolution”, Communications Biology, Vol.1, Article 84, https://doi.org/10.1038/s42003-018-0089-4.
Xie, E. et al. (2019), “A strategy for generating rice apomixis by gene editing”, Journal of Integrative Plant Biology, Vol. 61, pp. 911-916, https://doi.org/10.1111/jipb.12785.
Xie, F. and J. Zhang (2018), “Shanyou 63: An elite mega rice hybrid in China”, Rice, Vol. 11, Article 17, https://doi.org/10.1186/s12284-018-0210-9.
Xie, L. and J. Lin (1980), “Studies on rice bunchy stunt disease of rice, a new virus disease of rice plant”, Chinese Science Bulletin, Vol. 25, pp. 785-789, https://doi.org/10.1360/sb1980-25-9-785.
Xu, P. et al. (2014), “Mapping three new interspecific hybrid sterile loci between Oryza sativa and O. glaberrima”, Breeding Science, Vol. 63, pp. 476-482, https://doi.org/10.1270/jsbbs.63.476.
Xu, R. et al. (2016), “Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice”, Journal of Genetics and Genomics, Vol. 43, pp. 529-532, https://doi.org/10.1016/j.jgg.2016.07.003.
Xu, R. et al. (2014), “Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice”, Rice, Vol. 7, Article 5, https://doi.org/10.1186/s12284-014-0005-6.
Xu, Y.-B. and Z.-T. Shen (1987), “Inheritance of stigma exsertion in rice”, Rice Genetics Newsletter, Vol. 4, pp. 76-77, https://shigen.nig.ac.jp/rice/oryzabase/asset/rgn/vol4/v4p76.html.
Xue, W. et al. (2008), “Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice”, Nature Genetics, Vol. 40, pp. 761-767, https://doi.org/10.1038/ng.143.
Yamamoto, E. (2010), “Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice”, Molecular Genetics and Genomics, Vol. 283, pp. 305-315, https://doi.org/10.1007/s00438-010-0514-y.
Yamamoto, E. et al. (2007), “Interaction of two recessive genes, hbd2 and hbd3, induces hybrid breakdown in rice”, Theoretical and Applied Genetics, Vol. 115, pp. 187-194, https://doi.org/10.1007/s00122-007-0554-9.
Yamamoto, N. et al. (2018), “Comparative whole genome re-sequencing analysis in upland New Rice for Africa: Insights into the breeding history and respective genome compositions”, Rice, Vol.11, Article 33, https://doi.org/10.1186/s12284-018-0224-3.
Yamamoto, T. et al. (2010), “Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms”, BMC Genomics, Vol. 11, Article 267, https://doi.org/10.1186/1471-2164-11-267.
Yamamoto, T., J. Yonemaru and M. Yano (2009), “Towards the understanding of complex traits in rice: Substantially or superficially?”, DNA Research, Vol. 16, pp. 141-154, https://doi.org/10.1093/dnares/dsp006.
Yamauchi, T. et al. (2009), “Homologous recombination-mediated knock-in targeting of the MET1α gene for a maintenance DNA methyltransferase reproducibly reveals dosage-dependent spatiotemporal gene expression in rice”, The Plant Journal, Vol. 60, pp. 386-396, https://doi.org/10.1111/j.1365-313X.2009.03947.x.
Yan, W.G. and S.F. Li (1987), “Study on out-crossing characteristics among male sterile lines containing same nucleus in rice”, Hybrid Rice, Vol. 4, pp. 8-11.
Yan, W.G. et al. (2009), “Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.)”, Molecular Breeding, Vol 24, pp. 277-292, https://doi.org/10.1007/s11032-009-9290-y.
Yan, X. and N.J. Talbot (2016), “Investigating the cell biology of plant infection by the rice blast fungus Magnaporthe oryzae”, Current Opinion in Microbiology, Vol. 34, pp. 147-153, https://doi.org/10.1016/j.mib.2016.10.001.
Yanagihara, S., H. Kato and H. Ikehashi (1992), “A new locus for multiple alleles causing hybrid sterility between an Aus variety and Javanica varieties in rice (Oryza sativa L.)”, Japanese Journal of Breeding, Vol. 42, pp. 793-801, https://doi.org/10.1270/jsbbs1951.42.793.
Yang, J. et al. (2010), “A killer-protector system regulates both hybrid sterility and segregation distortion in rice”, Science, Vol. 337, pp. 1336-1340.
Yang, L. et al. (2006), “A transgenic rice seed accumulating an anti-hypertensive peptide reduces the blood pressure of spontaneously hypertensive rats”, FEBS Letters, Vol. 580, pp. 3315-3320, https://doi.org/10.1016/j.febslet.2006.04.092.
Yang, W., K.A. Weber and W.L. Silver (2012), “Nitrogen loss from soil through anaerobic ammonium oxidation coupled to iron reduction”, Nature Geoscience, Vol. 5, pp. 538-541, https://doi.org/10.1038/ngeo1530.
Yang, X. et al. (2017), “Knocking out of carotenoid catabolic genes in rice fails to boost carotenoid accumulation, but reveals a mutation in strigolactone biosynthesis”, Plant Cell Reports, Vol. 36, pp. 1533-1545, https://doi.org/10.1007/s00299-017-2172-6.
Yano, M. (2001), “Genetic and molecular dissection of naturally occurring variation”, Current Opinion in Plant Biology, Vol. 4, pp. 130-135, https://doi.org/10.1016/S1369-5266(00)00148-5.
Yano, M. et al. (2000), “Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS”, The Plant Cell, Vol. 12, pp. 2473-2483, https://doi.org/10.1105/tpc.12.12.2473.
Yano, M. et al. (2001), “Genetic control of flowering time in rice, a short-day plant”, Plant Physiology, Vol. 127, pp. 1425-1429, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1540174/.
Ye, X. et al. (2000), “Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm”, Science, Vol. 287, pp. 303-305, https://doi.org/10.1126/science.287.5451.303.
Yi, P. et al. (2002), “Discovery of mitochondrial chimeric-gene associated with cytoplasmic male sterility of HL-rice”, Chinese Science Bulletin, Vol. 47, pp. 744-747, https://doi.org/10.1360/02tb9168.
Yin, X. et al. (2017), “CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice”, Plant Cell Reports, Vol. 36, pp. 745-757, https://doi.org/10.1007/s00299-017-2118-z.
Yoshida, A. et al. (2016), “Analysis of rhizome development in Oryza longistaminata, a wild rice species”, Plant and Cell Physiology, Vol. 57, pp. 2213-2220, https://doi.org/10.1093/pcp/pcw138.
Yoshida, H. et al. (2007), “superwoman1-cleistogamy, a hopeful allele for gene containment in GM rice”, Plant Biotechnology Journal, Vol. 5, pp. 835-846, https://doi.org/10.1111/j.1467-7652.2007.00291.x.
Yoshida, S. (1981), “Heading and anthesis”, in Fundamentals of Rice Crop Science, International Rice Research Institute, Los Baños, Philippines, p. 55-58, http://books.irri.org/9711040522_content.pdf.
Yoshihara, T. et al. (1980), “Oxalic acid as a sucking inhibitor of the brown planthopper in rice (Delphacidae, Homoptera)”, Entomologia Exp. et Applicata, Vol. 27, pp. 149-155, https://doi.org/10.1111/j.1570-7458.1980.tb02959.x.
Yu, J. et al. (2005), “The genomes of Oryza sativa: A history of duplications”, PLoS Biology, Vol. 3, e38, https://doi.org/10.1371/journal.pbio.0030038.
Yu, J. et al. (2002), “A draft sequence of the rice genome (Oryza sativa L. ssp. indica)”, Science, Vol. 296, pp. 79-92, https://doi.org/10.1126/science.1068037.
Yu, Y. et al. (2016), “Hybrid sterility in rice (Oryza sativa L.) involves the tetratricopeptide repeat domain containing protein”, Genetics, Vol. 203, pp. 1439-1451, https://doi.org/10.1534/genetics.115.183848.
Zhang, H. et al. (2017), “Genome editing—principles and applications for functional genomics research and crop improvement”, Critical Reviews in Plant Sciences, Vol. 36, pp. 291-309, https://doi.org/10.1080/07352689.2017.1402989.
Zhang, H.-M. et al. (2008), “A black-streaked dwarf disease on rice in China is caused by a novel fijivirus”, Archives of Virology, Vol.153, pp.1893-1898, https://doi.org/10.1007/s00705-008-0209-4.
Zhang, H.M. et al. (1988), “Transgenic rice plants produced by electroporation-mediated plasmid uptake into protoplasts”, Plant Cell Reports, Vol. 7, pp. 379-384, https://doi.org/10.1007/BF00269517.
Zhang, J. et al. (2019), “NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice”, Nature Biotechnology, Vol. 37, pp. 676-684, https://doi.org/10.1038/s41587-019-0104-4.
Zhang, Q.J. et al. (2014), “Rapid diversification of five Oryza AA genomes associated with rice adaptation”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 111, pp. E4954-E4962, https://doi.org/10.1073/pnas.1418307111.
Zhang, W. and R. Wu (1988), “Efficient regeneration of transgenic plants from rice protoplasts and correctly regulated expression of the foreign gene in the plants”, Theoretical and Applied Genetics, Vol. 76, pp. 835-840, https://doi.org/10.1007/BF00273668.
Zhang, Y. et al. (2015), “Genome and comparative transcriptomics of African wild rice Oryza longistaminata provide insights into molecular mechanism of rhizomatousness and self-incompatibility”, Molecular Plant, Vol. 8, pp. 1683–1686, https://doi.org/10.1016/j.molp.2015.08.006.
Zhang, Y. et al. (2011), “Fine mapping of a gene responsible for pollen semi-sterility in hybrids between Oryza sativa L. and O. glaberrima Steud”, Molecular Breeding, Vol. 28, pp. 323-334, https://doi.org/10.1007/s11032-010-9485-2.
Zhang, Z. (1992), “Pest insects of rice in China”, in Z. Xiong et al. (eds.), Rice in China, China Agricultural Science and Technology Press, Beijing, pp. 130-149.
Zhang, Z. et al. (2002), “A new sterile gene from Oryza glaberrima on chromosome 3”, Rice Genetics Newsletter, Vol. 22, pp. 26-28, https://archive.gramene.org/newsletters/rice_genetics/rgn22/v05.html.
Zhao, Q. et al. (2018), “Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice”, Nature Genetics, Vol. 50, pp. 278-284, https://doi.org/10.1038/s41588-018-0041-z.
Zheng, Y. et al. (2016), “Rice domestication revealed by reduced shattering of archaeological rice from the lower Yangtze valley”, Scientific Reports, Vol. 6, Article 28136, https://doi.org/10.1038/srep28136.
Zhou, G. et al. (2013), “Southern rice black-streaked dwarf virus: A white-backed planthopper-transmitted fijivirus threatening rice production in Asia”, Frontiers in Microbiology, Vol. 4, 270, https://doi.org/10.3389/fmicb.2013.00270.
Zhou, G. et al. (2008), “Southern rice black-streaked dwarf virus: A new proposed Fijivirus species in the family Reoviridae”, Chinese Science Bulletin, Vol. 53, pp. 3677-3685, https://doi.org/10.1007/s11434-008-0467-2.
Zhou, G.W. et al. (2016), “Electron shuttles enhance anaerobic ammonium oxidation coupled to iron (III) reduction”, Environmental Science and Technology, Vol. 50, pp. 9298-9307, https://doi.org/10.1021/acs.est.6b02077.
Zhou, H. et al. (2016), “Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system”, Scientific Reports, Vol. 6, Article 37395, https://doi.org/10.1038/srep37395.
Zhou, J. et al. (2015), “Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice”, The Plant Journal, Vol. 82, pp. 632-643, https://doi.org/10.1111/tpj.12838.
Zhu, Y. et al. (2017), “Draft genome sequence of rice orange leaf phytoplasma from Guangdong, China”, Genome Announcements, Vol. 5/22, https://doi.org/10.1128/genomeA.00430-17.
Zhuang, C. et al. (2002), “Molecular mapping of S-c, an F1 pollen sterility gene in cultivated rice”, Euphytica, Vol. 127, pp. 133-138, https://doi.org/10.1023/A:1019973110467.
Zhuang, C. et al. (1999), “Molecular mapping of the S-a locus for F1 pollen sterility in cultivated rice (Oryza sativa L.)”, Acta Genetica Sinica, Vol. 26, pp. 213-218, https://europepmc.org/article/med/10589160.
Ziska, L.H. et al. (2015), “Weedy (red) rice: An emerging constraint to global rice production”, Advances in Agronomy, Vol. 129, pp. 181-228, https://doi.org/10.1016/bs.agron.2014.09.003.
Zong, Y. et al. (2017), “Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion”, Nature Biotechnology, Vol. 35, pp. 438-440, https://doi.org/10.1038/nbt.3811.
Zou, X.H. et al. (2015), “Multiple origins of BBCC allopolyploid species in the rice genus (Oryza)”, Scientific Reports, Vol. 5, Article 14876, https://doi.org/10.1038/srep14876.
Zuo, J. and J. Li (2014), “Molecular genetic dissection of quantitative trait loci regulating rice grain size”, Annual Review of Genetics, Vol. 48, pp. 99-118, https://doi.org/10.1146/annurev-genet-120213-092138.